| 1 |
|
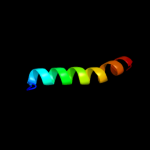
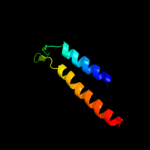
PDB 2y69 chain Z
Region: 37 - 63
Aligned: 27
Modelled: 27
Confidence: 48.1%
Identity: 30%
PDB header:electron transport
Chain: Z: PDB Molecule:cytochrome c oxidase polypeptide 8h;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 2 |
|
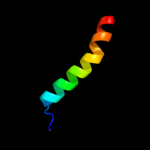
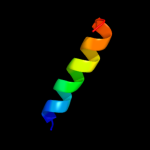
PDB 1v54 chain M
Region: 37 - 63
Aligned: 27
Modelled: 27
Confidence: 40.3%
Identity: 30%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Family: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Phyre2
| 3 |
|
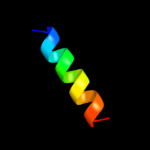
PDB 2jwa chain A
Region: 8 - 24
Aligned: 17
Modelled: 17
Confidence: 10.2%
Identity: 29%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 4 |
|
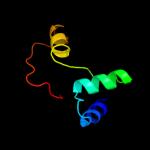
PDB 3fhg chain A
Region: 26 - 87
Aligned: 59
Modelled: 62
Confidence: 9.8%
Identity: 10%
PDB header:dna repair, hydrolase, lyase
Chain: A: PDB Molecule:n-glycosylase/dna lyase;
PDBTitle: crystal structure of sulfolobus solfataricus 8-oxoguanine dna2 glycosylase (ssogg)
Phyre2
| 5 |
|
PDB 3dww chain A
Region: 13 - 71
Aligned: 54
Modelled: 59
Confidence: 7.9%
Identity: 17%
PDB header:isomerase
Chain: A: PDB Molecule:prostaglandin e synthase;
PDBTitle: electron crystallographic structure of human microsomal2 prostaglandin e synthase 1
Phyre2
| 6 |
|
PDB 1wu0 chain A
Region: 13 - 31
Aligned: 19
Modelled: 19
Confidence: 6.0%
Identity: 5%
PDB header:hydrolase
Chain: A: PDB Molecule:atp synthase c chain;
PDBTitle: solution structure of subunit c of f1fo-atp synthase from2 the thermophilic bacillus ps3
Phyre2
| 7 |
|
PDB 2wpd chain P
Region: 13 - 29
Aligned: 17
Modelled: 17
Confidence: 5.8%
Identity: 29%
PDB header:hydrolase
Chain: P: PDB Molecule:atp synthase subunit 9, mitochondrial;
PDBTitle: the mg.adp inhibited state of the yeast f1c10 atp synthase
Phyre2