| 1 |
|
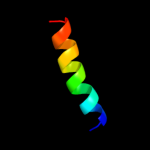
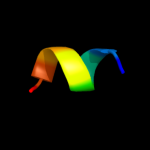
PDB 1w5c chain L
Region: 7 - 25
Aligned: 19
Modelled: 19
Confidence: 22.2%
Identity: 32%
PDB header:photosynthesis
Chain: L: PDB Molecule:cytochrome b559 beta subunit;
PDBTitle: photosystem ii from thermosynechococcus elongatus
Phyre2
| 2 |
|
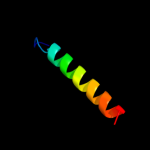
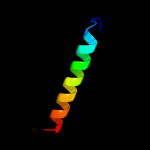
PDB 2axt chain F domain 1
Region: 7 - 25
Aligned: 19
Modelled: 19
Confidence: 21.5%
Identity: 32%
Fold: Single transmembrane helix
Superfamily: Cytochrome b559 subunits
Family: Cytochrome b559 subunits
Phyre2
| 3 |
|
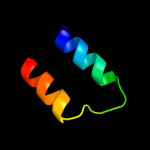
PDB 1c8s chain A
Region: 38 - 64
Aligned: 27
Modelled: 27
Confidence: 9.4%
Identity: 19%
Fold: Family A G protein-coupled receptor-like
Superfamily: Family A G protein-coupled receptor-like
Family: Bacteriorhodopsin-like
Phyre2
| 4 |
|
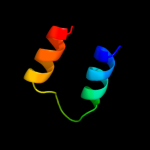
PDB 1v54 chain E
Region: 29 - 57
Aligned: 29
Modelled: 29
Confidence: 8.5%
Identity: 31%
Fold: alpha-alpha superhelix
Superfamily: Cytochrome c oxidase subunit E
Family: Cytochrome c oxidase subunit E
Phyre2
| 5 |
|
PDB 2y69 chain R
Region: 29 - 57
Aligned: 29
Modelled: 29
Confidence: 8.4%
Identity: 31%
PDB header:electron transport
Chain: R: PDB Molecule:cytochrome c oxidase subunit 5a;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 6 |
|
PDB 2k6t chain A
Region: 51 - 58
Aligned: 8
Modelled: 8
Confidence: 7.3%
Identity: 75%
PDB header:hormone
Chain: A: PDB Molecule:insulin-like 3 a chain;
PDBTitle: solution structure of the relaxin-like factor
Phyre2
| 7 |
|
PDB 2h8b chain A
Region: 51 - 58
Aligned: 8
Modelled: 8
Confidence: 7.3%
Identity: 75%
PDB header:hormone/growth factor
Chain: A: PDB Molecule:insulin-like 3;
PDBTitle: solution structure of insl3
Phyre2
| 8 |
|
PDB 2k6u chain A
Region: 51 - 58
Aligned: 8
Modelled: 8
Confidence: 7.2%
Identity: 75%
PDB header:hormone
Chain: A: PDB Molecule:insulin-like 3 a chain;
PDBTitle: the solution structure of a conformationally restricted2 fully active derivative of the human relaxin-like factor3 (rlf)
Phyre2
| 9 |
|
PDB 1m0k chain A
Region: 38 - 64
Aligned: 27
Modelled: 27
Confidence: 6.6%
Identity: 19%
PDB header:ion transport
Chain: A: PDB Molecule:bacteriorhodopsin;
PDBTitle: bacteriorhodopsin k intermediate at 1.43 a resolution
Phyre2