| 1 |
|
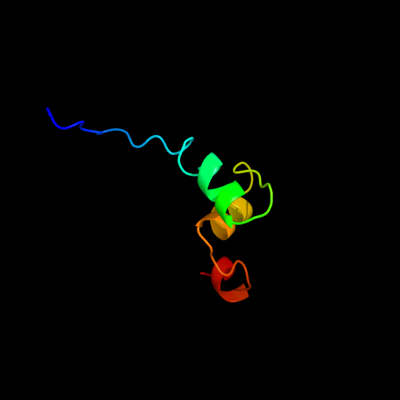
PDB 2gqb chain A domain 1
Region: 24 - 72
Aligned: 49
Modelled: 49
Confidence: 11.2%
Identity: 22%
Fold: RPA2825-like
Superfamily: RPA2825-like
Family: RPA2825-like
Phyre2
| 2 |
|
PDB 2v6z chain M
Region: 95 - 111
Aligned: 17
Modelled: 17
Confidence: 7.8%
Identity: 41%
PDB header:transferase
Chain: M: PDB Molecule:dna polymerase epsilon subunit 2;
PDBTitle: solution structure of amino terminal domain of human dna2 polymerase epsilon subunit b
Phyre2
| 3 |
|
PDB 3lyv chain F
Region: 119 - 130
Aligned: 12
Modelled: 12
Confidence: 7.3%
Identity: 17%
PDB header:chaperone
Chain: F: PDB Molecule:ribosome-associated factor y;
PDBTitle: crystal structure of a domain of ribosome-associated factor y from2 streptococcus pyogenes serotype m6. northeast structural genomics3 consortium target id dr64a
Phyre2
| 4 |
|
PDB 2h3o chain A
Region: 9 - 28
Aligned: 20
Modelled: 19
Confidence: 7.1%
Identity: 25%
PDB header:membrane protein
Chain: A: PDB Molecule:merf;
PDBTitle: structure of merft, a membrane protein with two trans-2 membrane helices
Phyre2
| 5 |
|
PDB 3haz chain A
Region: 46 - 106
Aligned: 59
Modelled: 61
Confidence: 6.5%
Identity: 15%
PDB header:oxidoreductase
Chain: A: PDB Molecule:proline dehydrogenase;
PDBTitle: crystal structure of bifunctional proline utilization a2 (puta) protein
Phyre2
| 6 |
|
PDB 3h3i chain A
Region: 81 - 98
Aligned: 18
Modelled: 18
Confidence: 6.1%
Identity: 28%
PDB header:lipid binding protein
Chain: A: PDB Molecule:putative lipid binding protein;
PDBTitle: crystal structure of a putative lipid binding protein (bt_2261) from2 bacteroides thetaiotaomicron vpi-5482 at 2.20 a resolution
Phyre2
| 7 |
|
PDB 1waz chain A
Region: 9 - 23
Aligned: 15
Modelled: 15
Confidence: 5.8%
Identity: 33%
PDB header:transport protein
Chain: A: PDB Molecule:merf;
PDBTitle: nmr structure determination of the bacterial mercury2 transporter, merf, in micelles
Phyre2