| 1 |
|
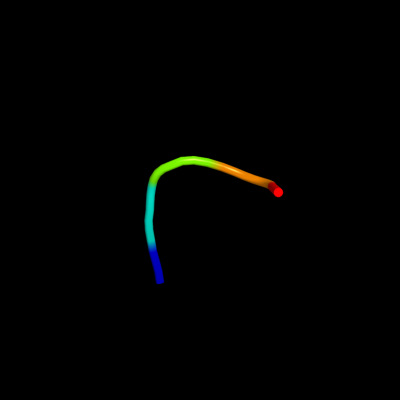
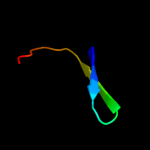
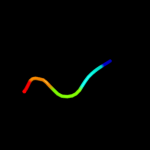
PDB 1wqk chain A
Region: 45 - 49
Aligned: 5
Modelled: 5
Confidence: 15.9%
Identity: 100%
PDB header:toxin
Chain: A: PDB Molecule:toxin apetx1;
PDBTitle: solution structure of apetx1, a specific peptide inhibitor2 of human ether-a-go-go-related gene potassium channels3 from the venom of the sea anemone anthopleura4 elegantissima: a new fold for an herg toxin
Phyre2
| 2 |
|
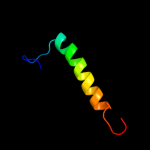
PDB 2guy chain A domain 1
Region: 9 - 19
Aligned: 11
Modelled: 11
Confidence: 11.3%
Identity: 27%
Fold: Glycosyl hydrolase domain
Superfamily: Glycosyl hydrolase domain
Family: alpha-Amylases, C-terminal beta-sheet domain
Phyre2
| 3 |
|
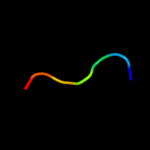
PDB 2aaa chain A domain 1
Region: 9 - 19
Aligned: 11
Modelled: 11
Confidence: 10.2%
Identity: 36%
Fold: Glycosyl hydrolase domain
Superfamily: Glycosyl hydrolase domain
Family: alpha-Amylases, C-terminal beta-sheet domain
Phyre2
| 4 |
|
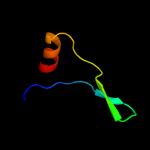
PDB 2taa chain A domain 1
Region: 9 - 19
Aligned: 11
Modelled: 11
Confidence: 8.7%
Identity: 27%
Fold: Glycosyl hydrolase domain
Superfamily: Glycosyl hydrolase domain
Family: alpha-Amylases, C-terminal beta-sheet domain
Phyre2
| 5 |
|
PDB 2qtx chain L
Region: 9 - 23
Aligned: 15
Modelled: 15
Confidence: 8.5%
Identity: 40%
PDB header:rna binding protein
Chain: L: PDB Molecule:uncharacterized protein mj1435;
PDBTitle: crystal structure of an hfq-like protein from methanococcus2 jannaschii
Phyre2
| 6 |
|
PDB 1kq1 chain W
Region: 9 - 28
Aligned: 20
Modelled: 20
Confidence: 7.0%
Identity: 20%
PDB header:translation
Chain: W: PDB Molecule:host factor for q beta;
PDBTitle: 1.55 a crystal structure of the pleiotropic translational2 regulator, hfq
Phyre2
| 7 |
|
PDB 1wxn chain A
Region: 45 - 49
Aligned: 5
Modelled: 5
Confidence: 6.4%
Identity: 60%
PDB header:toxin
Chain: A: PDB Molecule:toxin apetx2;
PDBTitle: solution structure of apetx2, a specific peptide inhibitor2 of asic3 proton-gated channels
Phyre2
| 8 |
|
PDB 2qsd chain B
Region: 11 - 51
Aligned: 41
Modelled: 41
Confidence: 6.3%
Identity: 27%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized conserved protein;
PDBTitle: crystal structure of a protein il1583 from idiomarina loihiensis
Phyre2
| 9 |
|
PDB 2hwn chain F
Region: 25 - 32
Aligned: 8
Modelled: 8
Confidence: 6.0%
Identity: 50%
PDB header:transferase
Chain: F: PDB Molecule:a kinase binding peptide;
PDBTitle: crystal structure of rii alpha dimerization/docking domain of pka2 bound to the d-akap2 peptide
Phyre2
| 10 |
|
PDB 2hwn chain E
Region: 25 - 32
Aligned: 8
Modelled: 8
Confidence: 6.0%
Identity: 50%
PDB header:transferase
Chain: E: PDB Molecule:a kinase binding peptide;
PDBTitle: crystal structure of rii alpha dimerization/docking domain of pka2 bound to the d-akap2 peptide
Phyre2
| 11 |
|
PDB 2qam chain C domain 1
Region: 42 - 51
Aligned: 10
Modelled: 10
Confidence: 6.0%
Identity: 40%
Fold: SH3-like barrel
Superfamily: Translation proteins SH3-like domain
Family: C-terminal domain of ribosomal protein L2
Phyre2
| 12 |
|
PDB 1p9q chain C domain 3
Region: 57 - 63
Aligned: 7
Modelled: 7
Confidence: 5.9%
Identity: 43%
Fold: Ferredoxin-like
Superfamily: EF-G C-terminal domain-like
Family: Hypothetical protein AF0491, C-terminal domain
Phyre2
| 13 |
|
PDB 1qf8 chain A
Region: 50 - 63
Aligned: 14
Modelled: 14
Confidence: 5.9%
Identity: 29%
Fold: Rubredoxin-like
Superfamily: Casein kinase II beta subunit
Family: Casein kinase II beta subunit
Phyre2
| 14 |
|
PDB 1u1s chain A domain 1
Region: 9 - 28
Aligned: 20
Modelled: 20
Confidence: 5.6%
Identity: 15%
Fold: Sm-like fold
Superfamily: Sm-like ribonucleoproteins
Family: Pleiotropic translational regulator Hfq
Phyre2
| 15 |
|
PDB 1t95 chain A domain 3
Region: 57 - 63
Aligned: 7
Modelled: 5
Confidence: 5.6%
Identity: 43%
Fold: Ferredoxin-like
Superfamily: EF-G C-terminal domain-like
Family: Hypothetical protein AF0491, C-terminal domain
Phyre2
| 16 |
|
PDB 1w7v chain D
Region: 12 - 44
Aligned: 33
Modelled: 33
Confidence: 5.4%
Identity: 18%
PDB header:hydrolase
Chain: D: PDB Molecule:xaa-pro aminopeptidase;
PDBTitle: znmg substituted aminopeptidase p from e. coli
Phyre2
| 17 |
|
PDB 2aiv chain A
Region: 44 - 50
Aligned: 7
Modelled: 7
Confidence: 5.4%
Identity: 100%
PDB header:transport protein
Chain: A: PDB Molecule:fragment of nucleoporin nup116/nsp116;
PDBTitle: multiple conformations in the ligand-binding site of the2 yeast nuclear pore targeting domain of nup116p
Phyre2
| 18 |
|
PDB 2guk chain A domain 1
Region: 26 - 33
Aligned: 8
Modelled: 8
Confidence: 5.2%
Identity: 13%
Fold: PG1857-like
Superfamily: PG1857-like
Family: PG1857-like
Phyre2
| 19 |
|
PDB 2i9x chain A domain 1
Region: 2 - 36
Aligned: 35
Modelled: 35
Confidence: 5.0%
Identity: 23%
Fold: SpoVG-like
Superfamily: SpoVG-like
Family: SpoVG-like
Phyre2