| 1 |
|
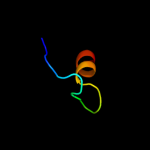
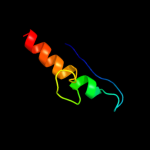
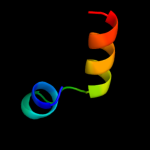
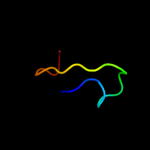
PDB 1o51 chain A
Region: 17 - 38
Aligned: 22
Modelled: 22
Confidence: 18.4%
Identity: 23%
Fold: Ferredoxin-like
Superfamily: GlnB-like
Family: DUF190/COG1993
Phyre2
| 2 |
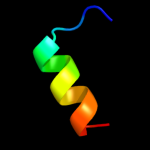
|
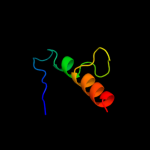
PDB 2dcl chain B
Region: 17 - 38
Aligned: 22
Modelled: 22
Confidence: 12.9%
Identity: 27%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical upf0166 protein ph1503;
PDBTitle: structure of ph1503 protein from pyrococcus horikoshii ot3
Phyre2
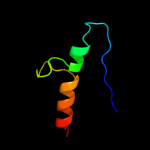
| 3 |
|
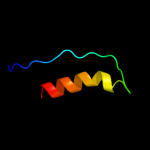
PDB 1bg2 chain A
Region: 2 - 43
Aligned: 42
Modelled: 42
Confidence: 11.1%
Identity: 21%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: Motor proteins
Phyre2
| 4 |
|
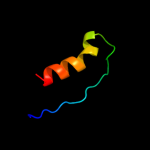
PDB 1qmy chain A
Region: 14 - 46
Aligned: 28
Modelled: 33
Confidence: 10.1%
Identity: 36%
Fold: Cysteine proteinases
Superfamily: Cysteine proteinases
Family: FMDV leader protease
Phyre2
| 5 |
|
PDB 2dlk chain A domain 1
Region: 8 - 23
Aligned: 14
Modelled: 16
Confidence: 9.9%
Identity: 57%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 6 |
|
PDB 1qol chain A
Region: 14 - 46
Aligned: 28
Modelled: 33
Confidence: 9.6%
Identity: 36%
Fold: Cysteine proteinases
Superfamily: Cysteine proteinases
Family: FMDV leader protease
Phyre2
| 7 |
|
PDB 3b6v chain A
Region: 2 - 43
Aligned: 42
Modelled: 42
Confidence: 7.4%
Identity: 17%
PDB header:motor protein
Chain: A: PDB Molecule:kinesin-like protein kif3c;
PDBTitle: crystal structure of the motor domain of human kinesin family member2 3c in complex with adp
Phyre2
| 8 |
|
PDB 3fbz chain A
Region: 22 - 43
Aligned: 22
Modelled: 22
Confidence: 6.3%
Identity: 45%
PDB header:structural protein
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of orf140 of the archaeal virus acidianus2 filamentous virus 1 (afv1)
Phyre2
| 9 |
|
PDB 1zfd chain A
Region: 6 - 16
Aligned: 11
Modelled: 11
Confidence: 6.1%
Identity: 27%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 10 |
|
PDB 3bex chain A domain 1
Region: 13 - 18
Aligned: 6
Modelled: 6
Confidence: 5.6%
Identity: 50%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: CoaX-like
Phyre2
| 11 |
|
PDB 2f9w chain A domain 2
Region: 13 - 18
Aligned: 6
Modelled: 6
Confidence: 5.6%
Identity: 50%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: CoaX-like
Phyre2
| 12 |
|
PDB 2dlk chain A domain 2
Region: 6 - 24
Aligned: 17
Modelled: 19
Confidence: 5.6%
Identity: 35%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 13 |
|
PDB 3fia chain A
Region: 29 - 43
Aligned: 15
Modelled: 15
Confidence: 5.6%
Identity: 47%
PDB header:protein binding
Chain: A: PDB Molecule:intersectin-1;
PDBTitle: crystal structure of the eh 1 domain from human intersectin-2 1 protein. northeast structural genomics consortium target3 hr3646e.
Phyre2
| 14 |
|
PDB 3cgu chain B
Region: 5 - 32
Aligned: 28
Modelled: 28
Confidence: 5.5%
Identity: 25%
PDB header:hormone/signaling protein
Chain: B: PDB Molecule:protein giant-lens;
PDBTitle: crystal structure of unliganded argos
Phyre2
| 15 |
|
PDB 1mkj chain A
Region: 2 - 43
Aligned: 42
Modelled: 42
Confidence: 5.3%
Identity: 21%
PDB header:transport protein
Chain: A: PDB Molecule:kinesin heavy chain;
PDBTitle: human kinesin motor domain with docked neck linker
Phyre2