| 1 |
|
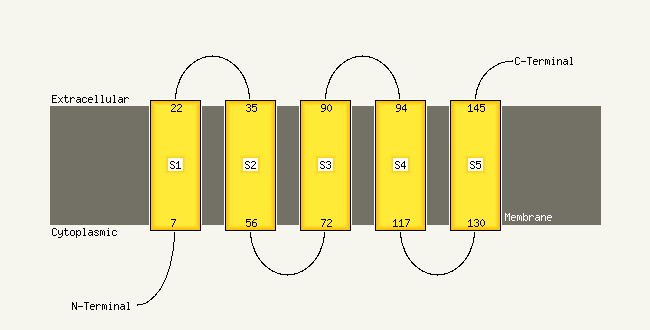
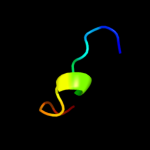
PDB 2knc chain A
Region: 67 - 91
Aligned: 25
Modelled: 25
Confidence: 41.5%
Identity: 16%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 2 |
|
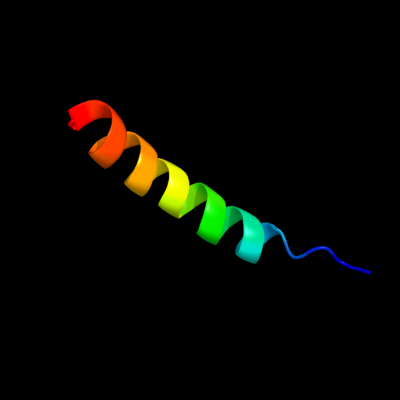
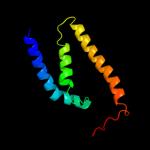
PDB 2b2h chain A
Region: 2 - 125
Aligned: 121
Modelled: 124
Confidence: 22.0%
Identity: 17%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
Phyre2
| 3 |
|
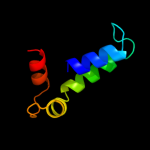
PDB 2d68 chain A
Region: 106 - 122
Aligned: 17
Modelled: 17
Confidence: 20.7%
Identity: 12%
PDB header:cell cycle
Chain: A: PDB Molecule:fop;
PDBTitle: structure of the n-terminal domain of fop (fgfr1op) protein
Phyre2
| 4 |
|
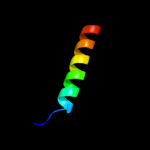
PDB 2e2z chain A
Region: 115 - 127
Aligned: 13
Modelled: 13
Confidence: 20.4%
Identity: 8%
PDB header:protein transport, chaperone regulator
Chain: A: PDB Molecule:tim15;
PDBTitle: solution nmr structure of yeast tim15, co-chaperone of2 mitochondrial hsp70
Phyre2
| 5 |
|
PDB 3hd6 chain A
Region: 33 - 126
Aligned: 94
Modelled: 94
Confidence: 17.7%
Identity: 16%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 6 |
|
PDB 2ptz chain A domain 2
Region: 20 - 80
Aligned: 53
Modelled: 61
Confidence: 14.6%
Identity: 15%
Fold: Enolase N-terminal domain-like
Superfamily: Enolase N-terminal domain-like
Family: Enolase N-terminal domain-like
Phyre2
| 7 |
|
PDB 2jp3 chain A
Region: 131 - 148
Aligned: 18
Modelled: 18
Confidence: 7.1%
Identity: 28%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 8 |
|
PDB 2k1a chain A
Region: 67 - 91
Aligned: 25
Modelled: 25
Confidence: 6.5%
Identity: 16%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
Phyre2