| 1 |
|
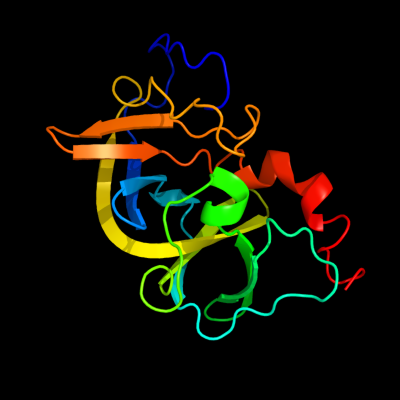
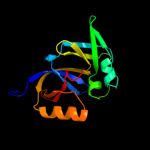
PDB 2ho9 chain A
Region: 1 - 167
Aligned: 167
Modelled: 167
Confidence: 100.0%
Identity: 100%
PDB header:signaling protein
Chain: A: PDB Molecule:chemotaxis protein chew;
PDBTitle: solution structure of chemotaxi protein chew from2 escherichia coli
Phyre2
| 2 |
|
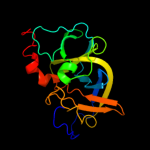
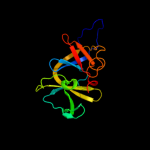
PDB 2qdl chain B
Region: 16 - 155
Aligned: 139
Modelled: 140
Confidence: 100.0%
Identity: 29%
PDB header:signaling protein
Chain: B: PDB Molecule:chemotaxis signal transduction protein;
PDBTitle: crystal structure of scaffolding protein ttchew from2 thermoanaerobacter tengcongensis
Phyre2
| 3 |
|
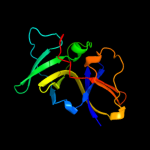
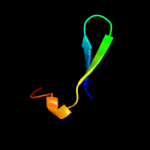
PDB 2ch4 chain W domain 1
Region: 16 - 153
Aligned: 135
Modelled: 138
Confidence: 100.0%
Identity: 24%
Fold: OB-fold
Superfamily: CheW-like
Family: CheW-like
Phyre2
| 4 |
|
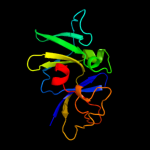
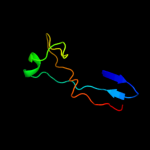
PDB 1b3q chain A domain 2
Region: 18 - 151
Aligned: 127
Modelled: 134
Confidence: 99.9%
Identity: 17%
Fold: OB-fold
Superfamily: CheW-like
Family: CheW-like
Phyre2
| 5 |
|
PDB 1b3q chain A
Region: 10 - 151
Aligned: 132
Modelled: 142
Confidence: 99.9%
Identity: 17%
PDB header:transferase
Chain: A: PDB Molecule:protein (chemotaxis protein chea);
PDBTitle: crystal structure of chea-289, a signal transducing histidine kinase
Phyre2
| 6 |
|
PDB 2ch4 chain A
Region: 3 - 151
Aligned: 139
Modelled: 149
Confidence: 99.7%
Identity: 17%
PDB header:transferase/chemotaxis
Chain: A: PDB Molecule:chemotaxis protein chea;
PDBTitle: complex between bacterial chemotaxis histidine kinase chea2 domains p4 and p5 and receptor-adaptor protein chew
Phyre2
| 7 |
|
PDB 2kk4 chain A
Region: 19 - 39
Aligned: 21
Modelled: 21
Confidence: 20.8%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein af_2094;
PDBTitle: solution nmr structure of protein af2094 from archaeoglobus2 fulgidus. northeast structural genomics consotium (nesg)3 target gt2
Phyre2
| 8 |
|
PDB 2xd0 chain B
Region: 20 - 70
Aligned: 50
Modelled: 51
Confidence: 11.9%
Identity: 16%
PDB header:toxin/rna
Chain: B: PDB Molecule:toxn;
PDBTitle: a processed non-coding rna regulates a bacterial antiviral2 system
Phyre2
| 9 |
|
PDB 2p12 chain A domain 1
Region: 129 - 167
Aligned: 39
Modelled: 39
Confidence: 8.4%
Identity: 26%
Fold: FomD barrel-like
Superfamily: FomD-like
Family: FomD-like
Phyre2