| 1 |
|
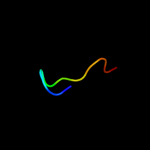
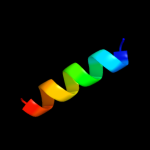
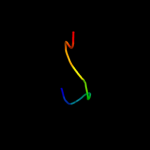
PDB 1xme chain C domain 1
Region: 6 - 14
Aligned: 9
Modelled: 9
Confidence: 12.5%
Identity: 67%
Fold: Single transmembrane helix
Superfamily: Bacterial ba3 type cytochrome c oxidase subunit IIa
Family: Bacterial ba3 type cytochrome c oxidase subunit IIa
Phyre2
| 2 |
|
PDB 2xfc chain D
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 12.3%
Identity: 45%
PDB header:virus
Chain: D: PDB Molecule:e1 envelope glycoprotein;
PDBTitle: the chikungunya e1 e2 envelope glycoprotein complex fit into2 the semliki forest virus cryo-em map
Phyre2
| 3 |
|
PDB 2xfb chain F
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 12.2%
Identity: 45%
PDB header:virus
Chain: F: PDB Molecule:e1 envelope glycoprotein;
PDBTitle: the chikungunya e1 e2 envelope glycoprotein complex fit into2 the sindbis virus cryo-em map
Phyre2
| 4 |
|
PDB 3n42 chain F
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 12.0%
Identity: 45%
PDB header:viral protein
Chain: F: PDB Molecule:e1 envelope glycoprotein;
PDBTitle: crystal structures of the mature envelope glycoprotein complex (furin2 cleavage) of chikungunya virus.
Phyre2
| 5 |
|
PDB 1z8y chain E
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 10.3%
Identity: 55%
PDB header:virus
Chain: E: PDB Molecule:spike glycoprotein e1;
PDBTitle: mapping the e2 glycoprotein of alphaviruses
Phyre2
| 6 |
|
PDB 3muw chain E
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 10.1%
Identity: 55%
PDB header:virus
Chain: E: PDB Molecule:structural polyprotein;
PDBTitle: pseudo-atomic structure of the e2-e1 protein shell in sindbis virus
Phyre2
| 7 |
|
PDB 1ld4 chain O
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 10.0%
Identity: 55%
PDB header:virus
Chain: O: PDB Molecule:spike glycoprotein e1;
PDBTitle: placement of the structural proteins in sindbis virus
Phyre2
| 8 |
|
PDB 2ala chain A domain 2
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 9.6%
Identity: 36%
Fold: Viral glycoprotein, central and dimerisation domains
Superfamily: Viral glycoprotein, central and dimerisation domains
Family: Viral glycoprotein, central and dimerisation domains
Phyre2
| 9 |
|
PDB 3j0c chain G
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 8.6%
Identity: 36%
PDB header:virus
Chain: G: PDB Molecule:e1 envelope glycoprotein;
PDBTitle: models of e1, e2 and cp of venezuelan equine encephalitis virus tc-832 strain restrained by a near atomic resolution cryo-em map
Phyre2
| 10 |
|
PDB 2ala chain A
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 8.4%
Identity: 36%
PDB header:viral protein
Chain: A: PDB Molecule:structural polyprotein (p130);
PDBTitle: crystal structure of the semliki forest virus envelope protein e1 in2 its monomeric conformation.
Phyre2
| 11 |
|
PDB 1q90 chain A domain 3
Region: 7 - 22
Aligned: 16
Modelled: 16
Confidence: 8.3%
Identity: 50%
Fold: Single transmembrane helix
Superfamily: Cytochrome f subunit of the cytochrome b6f complex, transmembrane anchor
Family: Cytochrome f subunit of the cytochrome b6f complex, transmembrane anchor
Phyre2
| 12 |
|
PDB 3muu chain A
Region: 41 - 51
Aligned: 11
Modelled: 11
Confidence: 6.3%
Identity: 55%
PDB header:viral protein
Chain: A: PDB Molecule:structural polyprotein;
PDBTitle: crystal structure of the sindbis virus e2-e1 heterodimer at low ph
Phyre2