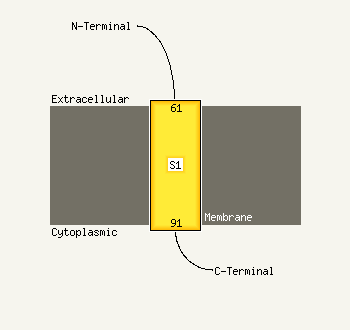
1 d1jkxa_
27.5
9
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase2 c2eenA_
25.4
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ph1819;PDBTitle: structure of ph1819 protein from pyrococcus horikoshii ot3
3 d1fmta2
21.8
10
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase4 c2zfdB_
17.2
11
PDB header: signaling protein/transferaseChain: B: PDB Molecule: putative uncharacterized protein t20l15_90;PDBTitle: the crystal structure of plant specific calcium binding protein atcbl22 in complex with the regulatory domain of atcipk14
5 d2bw0a2
17.2
8
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase6 c2v4oB_
17.0
21
PDB header: hydrolaseChain: B: PDB Molecule: multifunctional protein sur e;PDBTitle: crystal structure of salmonella typhimurium sure at 2.752 angstrom resolution in monoclinic form
7 c3oquB_
17.0
19
PDB header: hormone receptorChain: B: PDB Molecule: abscisic acid receptor pyl9;PDBTitle: crystal structure of native abscisic acid receptor pyl9 with aba
8 d1wjwa_
16.4
10
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain9 d2blna2
15.2
14
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase10 d2ae8a2
12.7
18
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase11 d1civa2
11.8
10
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain12 c2fjtA_
11.4
17
PDB header: lyaseChain: A: PDB Molecule: adenylyl cyclase class iv;PDBTitle: adenylyl cyclase class iv from yersinia pestis
13 d1xo8a_
11.3
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: LEA14-likeFamily: LEA14-like14 c3rfoA_
10.7
13
PDB header: transferaseChain: A: PDB Molecule: methionyl-trna formyltransferase;PDBTitle: crystal structure of methyionyl-trna formyltransferase from bacillus2 anthracis
15 c3p9xB_
9.9
21
PDB header: transferaseChain: B: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
16 d2ldxa2
9.3
8
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain17 d1ev0a_
8.8
27
Fold: Cell division protein MinE topological specificity domainSuperfamily: Cell division protein MinE topological specificity domainFamily: Cell division protein MinE topological specificity domain18 d2f1da2
8.6
21
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase19 d1s3ia2
8.6
10
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase20 c1ybxA_
8.5
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: conserved hypothetical protein cth-383 from clostridium thermocellum
21 c2y94C_
not modelled
8.4
36
PDB header: transferaseChain: C: PDB Molecule: 5'-amp-activated protein kinase catalytic subunit alpha-1;PDBTitle: structure of an active form of mammalian ampk
22 d1gsoa1
not modelled
8.1
6
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: BC C-terminal domain-like23 c2f1dP_
not modelled
7.6
21
PDB header: lyaseChain: P: PDB Molecule: imidazoleglycerol-phosphate dehydratase 1;PDBTitle: x-ray structure of imidazoleglycerol-phosphate dehydratase
24 c3n5fB_
not modelled
7.5
12
PDB header: hydrolaseChain: B: PDB Molecule: n-carbamoyl-l-amino acid hydrolase;PDBTitle: crystal structure of l-n-carbamoylase from geobacillus2 stearothermophilus cect43
25 d1rhya2
not modelled
7.4
11
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase26 c1rhyB_
not modelled
7.3
12
PDB header: lyaseChain: B: PDB Molecule: imidazole glycerol phosphate dehydratase;PDBTitle: crystal structure of imidazole glycerol phosphate dehydratase
27 c2ae8C_
not modelled
7.2
19
PDB header: lyaseChain: C: PDB Molecule: imidazoleglycerol-phosphate dehydratase;PDBTitle: crystal structure of imidazoleglycerol-phosphate dehydratase from2 staphylococcus aureus subsp. aureus n315
28 d2cmda2
not modelled
7.0
16
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain29 c1s1hJ_
not modelled
7.0
11
PDB header: ribosomeChain: J: PDB Molecule: 40s ribosomal protein s20;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
30 c2i3fA_
not modelled
6.7
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: glycolipid transfer-like protein;PDBTitle: crystal structure of a glycolipid transfer-like protein2 from galdieria sulphuraria
31 c2dc4A_
not modelled
6.7
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 165aa long hypothetical protein;PDBTitle: structure of ph1012 protein from pyrococcus horikoshii ot3
32 c3dcjA_
not modelled
6.5
14
PDB header: transferaseChain: A: PDB Molecule: probable 5'-phosphoribosylglycinamidePDBTitle: crystal structure of glycinamide formyltransferase (purn)2 from mycobacterium tuberculosis in complex with 5-methyl-5,3 6,7,8-tetrahydrofolic acid derivative
33 c2hjqA_
not modelled
6.5
14
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical protein yqbf;PDBTitle: nmr structure of bacillus subtilis protein yqbf, northeast2 structural genomics target sr449
34 d1a6da3
not modelled
6.4
10
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: Group II chaperonin (CCT, TRIC), intermediate domain35 c1yrwA_
not modelled
5.4
14
PDB header: transferaseChain: A: PDB Molecule: protein arna;PDBTitle: crystal structure of e.coli arna transformylase domain
36 d1q3qa3
not modelled
5.4
14
Fold: GroEL-intermediate domain likeSuperfamily: GroEL-intermediate domain likeFamily: Group II chaperonin (CCT, TRIC), intermediate domain