| 1 |
|
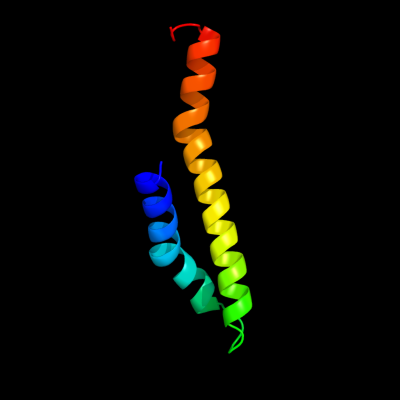
PDB 2oar chain A domain 1
Region: 132 - 197
Aligned: 66
Modelled: 66
Confidence: 41.3%
Identity: 15%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 2 |
|
PDB 2in1 chain A domain 1
Region: 119 - 148
Aligned: 30
Modelled: 30
Confidence: 35.8%
Identity: 23%
Fold: UBC-like
Superfamily: UBC-like
Family: UFC1-like
Phyre2
| 3 |
|
PDB 2ht2 chain B
Region: 3 - 134
Aligned: 86
Modelled: 86
Confidence: 26.3%
Identity: 13%
PDB header:membrane protein
Chain: B: PDB Molecule:h(+)/cl(-) exchange transporter clca;
PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
Phyre2
| 4 |
|
PDB 3kpa chain B
Region: 119 - 156
Aligned: 38
Modelled: 38
Confidence: 26.1%
Identity: 18%
PDB header:ligase
Chain: B: PDB Molecule:probable ubiquitin fold modifier conjugating enzyme;
PDBTitle: ubiquitin fold modifier conjugating enzyme from leishmania major2 (probable)
Phyre2
| 5 |
|
PDB 3nke chain A
Region: 127 - 184
Aligned: 58
Modelled: 58
Confidence: 23.3%
Identity: 21%
PDB header:immune system
Chain: A: PDB Molecule:protein ygbt;
PDBTitle: high resolution structure of the c-terminal domain crisp-associated2 protein cas1 from escherichia coli str. k-12
Phyre2
| 6 |
|
PDB 1xoo chain A
Region: 25 - 39
Aligned: 15
Modelled: 15
Confidence: 19.5%
Identity: 27%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin;
PDBTitle: nmr structure of g1s mutant of influenza hemagglutinin2 fusion peptide in dpc micelles at ph 5
Phyre2
| 7 |
|
PDB 1xop chain A
Region: 22 - 39
Aligned: 18
Modelled: 18
Confidence: 18.7%
Identity: 22%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin;
PDBTitle: nmr structure of g1v mutant of influenza hemagglutinin2 fusion peptide in dpc micelles at ph 5
Phyre2
| 8 |
|
PDB 1ibo chain A
Region: 25 - 39
Aligned: 15
Modelled: 15
Confidence: 18.1%
Identity: 27%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin ha2 chain peptide;
PDBTitle: nmr structure of hemagglutinin fusion peptide in dpc2 micelles at ph 7.4
Phyre2
| 9 |
|
PDB 1ibn chain A
Region: 25 - 39
Aligned: 15
Modelled: 15
Confidence: 18.1%
Identity: 27%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin ha2 chain peptide;
PDBTitle: nmr structure of hemagglutinin fusion peptide in dpc2 micelles at ph 5
Phyre2
| 10 |
|
PDB 2oar chain A
Region: 132 - 199
Aligned: 68
Modelled: 68
Confidence: 14.3%
Identity: 16%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 11 |
|
PDB 1ots chain A
Region: 2 - 134
Aligned: 87
Modelled: 87
Confidence: 12.7%
Identity: 13%
Fold: Clc chloride channel
Superfamily: Clc chloride channel
Family: Clc chloride channel
Phyre2
| 12 |
|
PDB 3b9y chain A
Region: 13 - 30
Aligned: 18
Modelled: 18
Confidence: 11.0%
Identity: 33%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 13 |
|
PDB 3god chain A
Region: 127 - 154
Aligned: 28
Modelled: 28
Confidence: 10.1%
Identity: 11%
PDB header:immune system
Chain: A: PDB Molecule:cas1;
PDBTitle: structural basis for dnase activity of a conserved protein2 implicated in crispr-mediated antiviral defense
Phyre2
| 14 |
|
PDB 2jrd chain A
Region: 25 - 39
Aligned: 15
Modelled: 15
Confidence: 10.0%
Identity: 20%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin;
PDBTitle: influenza hemagglutinin fusion domain mutant f9a
Phyre2
| 15 |
|
PDB 2kpe chain B
Region: 14 - 31
Aligned: 18
Modelled: 18
Confidence: 9.9%
Identity: 28%
PDB header:membrane protein
Chain: B: PDB Molecule:glycophorin-a;
PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
Phyre2
| 16 |
|
PDB 2kpe chain A
Region: 14 - 31
Aligned: 18
Modelled: 18
Confidence: 9.9%
Identity: 28%
PDB header:membrane protein
Chain: A: PDB Molecule:glycophorin-a;
PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
Phyre2
| 17 |
|
PDB 1kpl chain A
Region: 16 - 134
Aligned: 73
Modelled: 73
Confidence: 8.7%
Identity: 15%
Fold: Clc chloride channel
Superfamily: Clc chloride channel
Family: Clc chloride channel
Phyre2
| 18 |
|
PDB 2zt9 chain E
Region: 16 - 30
Aligned: 15
Modelled: 15
Confidence: 7.2%
Identity: 40%
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex from nostoc sp. pcc2 7120
Phyre2
| 19 |
|
PDB 3nkd chain B
Region: 127 - 170
Aligned: 44
Modelled: 44
Confidence: 6.3%
Identity: 16%
PDB header:immune system
Chain: B: PDB Molecule:crispr-associated protein cas1;
PDBTitle: structure of crisp-associated protein cas1 from escherichia coli str.2 k-12
Phyre2
| 20 |
|
PDB 1jeg chain B
Region: 38 - 53
Aligned: 16
Modelled: 15
Confidence: 5.8%
Identity: 31%
PDB header:transferase/hydrolase
Chain: B: PDB Molecule:hematopoietic cell protein-tyrosine phosphatase
PDBTitle: solution structure of the sh3 domain from c-terminal src2 kinase complexed with a peptide from the tyrosine3 phosphatase pep
Phyre2