| 1 |
|
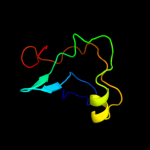
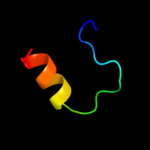
PDB 1z8g chain A domain 2
Region: 25 - 73
Aligned: 47
Modelled: 49
Confidence: 47.5%
Identity: 13%
Fold: SRCR-like
Superfamily: SRCR-like
Family: Hepsin, N-terminal domain
Phyre2
| 2 |
|
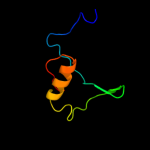
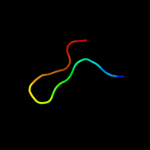
PDB 1z8g chain A
Region: 34 - 72
Aligned: 39
Modelled: 39
Confidence: 45.7%
Identity: 18%
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:serine protease hepsin;
PDBTitle: crystal structure of the extracellular region of the transmembrane2 serine protease hepsin with covalently bound preferred substrate.
Phyre2
| 3 |
|
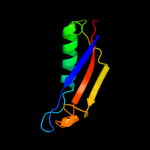
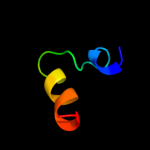
PDB 2ott chain Y
Region: 24 - 72
Aligned: 43
Modelled: 49
Confidence: 41.6%
Identity: 14%
PDB header:immune system
Chain: Y: PDB Molecule:t-cell surface glycoprotein cd5;
PDBTitle: crystal structure of cd5_diii
Phyre2
| 4 |
|
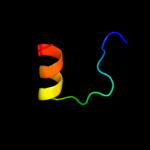
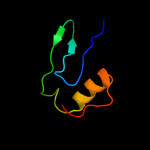
PDB 2jop chain A
Region: 24 - 87
Aligned: 64
Modelled: 64
Confidence: 26.6%
Identity: 9%
PDB header:immune system
Chain: A: PDB Molecule:t-cell surface glycoprotein cd5;
PDBTitle: solution structure of the n-terminal extracellular domain2 of the lymphocyte receptor cd5 (cd5 domain 1)
Phyre2
| 5 |
|
PDB 2oya chain A
Region: 18 - 72
Aligned: 52
Modelled: 55
Confidence: 24.4%
Identity: 13%
PDB header:ligand binding protein
Chain: A: PDB Molecule:macrophage receptor marco;
PDBTitle: crystal structure analysis of the dimeric form of the srcr domain of2 mouse marco
Phyre2
| 6 |
|
PDB 1dx5 chain I domain 1
Region: 95 - 100
Aligned: 6
Modelled: 6
Confidence: 14.4%
Identity: 50%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: EGF/Laminin
Family: EGF-type module
Phyre2
| 7 |
|
PDB 2y1b chain A
Region: 32 - 105
Aligned: 64
Modelled: 74
Confidence: 14.1%
Identity: 19%
PDB header:membrane protein
Chain: A: PDB Molecule:putative outer membrane protein, signal;
PDBTitle: crystal structure of the e. coli outer membrane lipoprotein2 rcsf
Phyre2
| 8 |
|
PDB 1ji8 chain A
Region: 39 - 59
Aligned: 21
Modelled: 21
Confidence: 13.5%
Identity: 24%
Fold: DsrC, the gamma subunit of dissimilatory sulfite reductase
Superfamily: DsrC, the gamma subunit of dissimilatory sulfite reductase
Family: DsrC, the gamma subunit of dissimilatory sulfite reductase
Phyre2
| 9 |
|
PDB 1yx3 chain A
Region: 39 - 59
Aligned: 21
Modelled: 21
Confidence: 11.9%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein dsrc;
PDBTitle: nmr structure of allochromatium vinosum dsrc: northeast2 structural genomics consortium target op4
Phyre2
| 10 |
|
PDB 1a0e chain A
Region: 64 - 77
Aligned: 14
Modelled: 14
Confidence: 9.4%
Identity: 29%
Fold: TIM beta/alpha-barrel
Superfamily: Xylose isomerase-like
Family: Xylose isomerase
Phyre2
| 11 |
|
PDB 1fvi chain A domain 1
Region: 91 - 107
Aligned: 16
Modelled: 17
Confidence: 9.3%
Identity: 31%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: DNA ligase/mRNA capping enzyme postcatalytic domain
Phyre2
| 12 |
|
PDB 2v4j chain C domain 1
Region: 39 - 60
Aligned: 22
Modelled: 22
Confidence: 8.8%
Identity: 9%
Fold: DsrC, the gamma subunit of dissimilatory sulfite reductase
Superfamily: DsrC, the gamma subunit of dissimilatory sulfite reductase
Family: DsrC, the gamma subunit of dissimilatory sulfite reductase
Phyre2
| 13 |
|
PDB 1a0c chain A
Region: 64 - 77
Aligned: 14
Modelled: 14
Confidence: 8.5%
Identity: 14%
Fold: TIM beta/alpha-barrel
Superfamily: Xylose isomerase-like
Family: Xylose isomerase
Phyre2
| 14 |
|
PDB 2a5w chain C
Region: 39 - 57
Aligned: 19
Modelled: 19
Confidence: 8.4%
Identity: 32%
PDB header:oxidoreductase
Chain: C: PDB Molecule:sulfite reductase, desulfoviridin-type subunit gamma
PDBTitle: crystal structure of the oxidized gamma-subunit of the dissimilatory2 sulfite reductase (dsrc) from archaeoglobus fulgidus
Phyre2
| 15 |
|
PDB 1by2 chain A
Region: 24 - 72
Aligned: 47
Modelled: 49
Confidence: 7.5%
Identity: 15%
Fold: SRCR-like
Superfamily: SRCR-like
Family: Scavenger receptor cysteine-rich (SRCR) domain
Phyre2
| 16 |
|
PDB 1xiw chain B
Region: 96 - 106
Aligned: 11
Modelled: 11
Confidence: 7.4%
Identity: 36%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: I set domains
Phyre2
| 17 |
|
PDB 1xzp chain A domain 3
Region: 93 - 101
Aligned: 9
Modelled: 9
Confidence: 6.6%
Identity: 22%
Fold: Folate-binding domain
Superfamily: Folate-binding domain
Family: TrmE formyl-THF-binding domain
Phyre2
| 18 |
|
PDB 1xzq chain B
Region: 93 - 101
Aligned: 9
Modelled: 9
Confidence: 6.6%
Identity: 22%
PDB header:hydrolase
Chain: B: PDB Molecule:probable trna modification gtpase trme;
PDBTitle: structure of the gtp-binding protein trme from thermotoga2 maritima complexed with 5-formyl-thf
Phyre2
| 19 |
|
PDB 2j8j chain A
Region: 31 - 79
Aligned: 49
Modelled: 49
Confidence: 6.5%
Identity: 16%
PDB header:hydrolase
Chain: A: PDB Molecule:coagulation factor xi;
PDBTitle: solution structure of the a4 domain of blood coagulation2 factor xi
Phyre2
| 20 |
|
PDB 1sy6 chain A domain 1
Region: 96 - 101
Aligned: 6
Modelled: 6
Confidence: 6.4%
Identity: 67%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: I set domains
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|