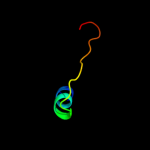
1 c1qp6B_
30.7
50
PDB header: de novo proteinChain: B: PDB Molecule: protein (alpha2d);PDBTitle: solution structure of alpha2d
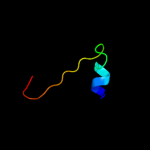
2 d1xfja_
23.3
27
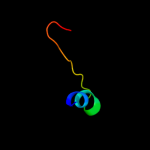
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: YfiH-like3 d1rw0a_
19.8
27
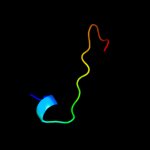
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: YfiH-like4 d2a6qb1
17.8
14
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like5 d1xafa_
17.5
33
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: YfiH-like6 d1t8ha_
16.6
27
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: YfiH-like7 c3hs2H_
16.3
14
PDB header: antitoxinChain: H: PDB Molecule: prevent host death protein;PDBTitle: crystal structure of phd truncated to residue 57 in an orthorhombic2 space group
8 c3hryA_
16.3
14
PDB header: antitoxinChain: A: PDB Molecule: prevent host death protein;PDBTitle: crystal structure of phd in a trigonal space group and partially2 disordered
9 d1bxna1
13.8
9
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain10 d1ykwa1
13.2
32
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain11 d1rv9a_
12.9
20
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: YfiH-like12 c3g5oA_
11.1
11
PDB header: toxin/antitoxinChain: A: PDB Molecule: uncharacterized protein rv2865;PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
13 c2ip6A_
10.9
33
PDB header: antimicrobial proteinChain: A: PDB Molecule: papb;PDBTitle: crystal structure of pedb
14 c2zt9E_
10.8
50
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from nostoc sp. pcc2 7120
15 c2j5dA_
9.6
40
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: nmr structure of bnip3 transmembrane domain in lipid2 bicelles
16 c2jobA_
9.3
30
PDB header: lipid binding proteinChain: A: PDB Molecule: antilipopolysaccharide factor;PDBTitle: solution structure of an antilipopolysaccharide factor from2 shrimp and its possible lipid a binding site
17 c3fy6A_
9.2
57
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: integron cassette protein;PDBTitle: structure from the mobile metagenome of v. cholerae.2 integron cassette protein vch_cass3
18 d1ldda_
8.8
0
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: SCF ubiquitin ligase complex WHB domain19 d1wwca_
8.6
25
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains20 c1rcxH_
8.2
32
PDB header: lyase (carbon-carbon)Chain: H: PDB Molecule: ribulose bisphosphate carboxylase/oxygenase;PDBTitle: non-activated spinach rubisco in complex with its substrate2 ribulose-1,5-bisphosphate
21 d2q9oa1
not modelled
8.1
42
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins22 d2a6qa1
not modelled
7.9
11
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like23 c1bwvA_
not modelled
7.6
14
PDB header: lyaseChain: A: PDB Molecule: protein (ribulose bisphosphate carboxylase);PDBTitle: activated ribulose 1,5-bisphosphate carboxylase/oxygenase (rubisco)2 complexed with the reaction intermediate analogue 2-carboxyarabinitol3 1,5-bisphosphate
24 d1vcaa2
not modelled
7.3
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains25 d1geha1
not modelled
6.8
27
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain26 d1bwva1
not modelled
6.7
18
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain27 d8ruca1
not modelled
6.6
32
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain28 d1wdda1
not modelled
6.5
32
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain29 c2jmvA_
not modelled
6.5
41
PDB header: antiviral proteinChain: A: PDB Molecule: scytovirin;PDBTitle: solution structure of scytovirin refined against residual2 dipolar couplings
30 d1ej7l1
not modelled
6.2
32
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain31 d1svda1
not modelled
6.2
27
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain32 c1vf5R_
not modelled
6.2
16
PDB header: photosynthesisChain: R: PDB Molecule: protein pet l;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
33 c2e75E_
not modelled
6.2
16
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
34 c2e74E_
not modelled
6.2
16
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
35 d2e74e1
not modelled
6.2
16
Fold: Single transmembrane helixSuperfamily: PetL subunit of the cytochrome b6f complexFamily: PetL subunit of the cytochrome b6f complex36 c2e76E_
not modelled
6.2
16
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
37 d1wwwx_
not modelled
6.1
26
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains38 c1telA_
not modelled
6.0
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ribulose bisphosphate carboxylase, large subunit;PDBTitle: crystal structure of a rubisco-like protein from chlorobium2 tepidum
39 c2w0cR_
not modelled
5.9
31
PDB header: virusChain: R: PDB Molecule: protein p3;PDBTitle: x-ray structure of the entire lipid-containing2 bacteriophage pm2
40 c3nwrA_
not modelled
5.9
32
PDB header: lyaseChain: A: PDB Molecule: a rubisco-like protein;PDBTitle: crystal structure of a rubisco-like protein from burkholderia fungorum
41 d1kbva1
not modelled
5.8
33
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins42 c2qygC_
not modelled
5.7
32
PDB header: unknown functionChain: C: PDB Molecule: ribulose bisphosphate carboxylase-like protein 2;PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
43 c2rq2A_
not modelled
5.6
45
PDB header: antibioticChain: A: PDB Molecule: big defensin;PDBTitle: the solution structure of the n-terminal fragment of big2 defensin