| 1 |
|
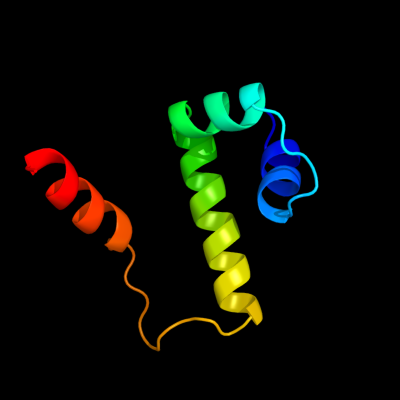
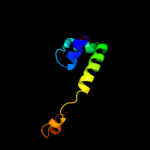
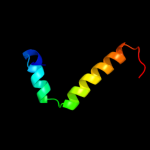
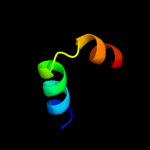
PDB 1or7 chain C
Region: 1 - 66
Aligned: 66
Modelled: 66
Confidence: 100.0%
Identity: 100%
PDB header:transcription
Chain: C: PDB Molecule:sigma-e factor negative regulatory protein;
PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
Phyre2
| 2 |
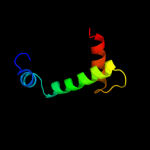
|
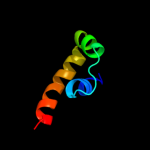
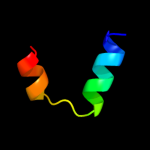
PDB 1or7 chain C
Region: 1 - 66
Aligned: 66
Modelled: 66
Confidence: 100.0%
Identity: 100%
Fold: N-terminal, cytoplasmic domain of anti-sigmaE factor RseA
Superfamily: N-terminal, cytoplasmic domain of anti-sigmaE factor RseA
Family: N-terminal, cytoplasmic domain of anti-sigmaE factor RseA
Phyre2
| 3 |
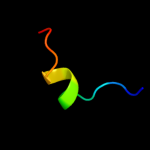
|
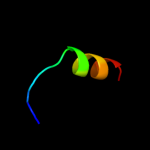
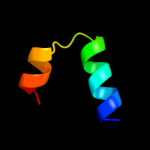
PDB 3m4w chain H
Region: 134 - 188
Aligned: 31
Modelled: 31
Confidence: 97.3%
Identity: 100%
PDB header:signaling protein/signaling protein
Chain: H: PDB Molecule:sigma-e factor negative regulatory protein;
PDBTitle: structural basis for the negative regulation of bacterial stress2 response by rseb
Phyre2
| 4 |
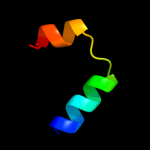
|
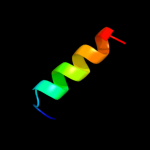
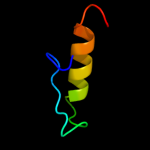
PDB 2z2s chain D
Region: 2 - 67
Aligned: 66
Modelled: 65
Confidence: 95.0%
Identity: 15%
PDB header:transcription
Chain: D: PDB Molecule:anti-sigma factor chrr, transcriptional activator chrr;
PDBTitle: crystal structure of rhodobacter sphaeroides sige in complex with the2 anti-sigma chrr
Phyre2
| 5 |
|
PDB 3hug chain J
Region: 3 - 45
Aligned: 43
Modelled: 43
Confidence: 94.8%
Identity: 16%
PDB header:transcription/membrane protein
Chain: J: PDB Molecule:probable conserved membrane protein;
PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
Phyre2
| 6 |
|
PDB 1t98 chain A domain 1
Region: 10 - 25
Aligned: 16
Modelled: 16
Confidence: 25.5%
Identity: 38%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: MukF N-terminal domain-like
Phyre2
| 7 |
|
PDB 1u04 chain A domain 1
Region: 23 - 40
Aligned: 18
Modelled: 18
Confidence: 19.8%
Identity: 50%
Fold: SH3-like barrel
Superfamily: PAZ domain
Family: PAZ domain
Phyre2
| 8 |
|
PDB 1m98 chain A domain 1
Region: 18 - 67
Aligned: 50
Modelled: 50
Confidence: 9.9%
Identity: 10%
Fold: Orange carotenoid protein, N-terminal domain
Superfamily: Orange carotenoid protein, N-terminal domain
Family: Orange carotenoid protein, N-terminal domain
Phyre2
| 9 |
|
PDB 1jhg chain A
Region: 8 - 49
Aligned: 42
Modelled: 42
Confidence: 7.3%
Identity: 19%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: TrpR-like
Family: Trp repressor, TrpR
Phyre2
| 10 |
|
PDB 1trr chain A
Region: 10 - 67
Aligned: 58
Modelled: 58
Confidence: 7.1%
Identity: 16%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: TrpR-like
Family: Trp repressor, TrpR
Phyre2
| 11 |
|
PDB 1yfn chain G
Region: 77 - 91
Aligned: 15
Modelled: 15
Confidence: 6.5%
Identity: 100%
PDB header:protein binding
Chain: G: PDB Molecule:sigma-e factor negative regulatory protein;
PDBTitle: versatile modes of peptide recognition by the aaa+ adaptor2 protein sspb- the crystal structure of a sspb-rsea complex
Phyre2
| 12 |
|
PDB 1g2y chain C
Region: 1 - 23
Aligned: 23
Modelled: 23
Confidence: 6.3%
Identity: 30%
PDB header:transcription
Chain: C: PDB Molecule:hepatocyte nuclear factor 1-alpha;
PDBTitle: hnf-1alpha dimerization domain, with selenomethionine2 substitued at leu 12
Phyre2
| 13 |
|
PDB 1g2y chain D
Region: 1 - 23
Aligned: 23
Modelled: 23
Confidence: 6.0%
Identity: 30%
PDB header:transcription
Chain: D: PDB Molecule:hepatocyte nuclear factor 1-alpha;
PDBTitle: hnf-1alpha dimerization domain, with selenomethionine2 substitued at leu 12
Phyre2
| 14 |
|
PDB 1g2y chain B
Region: 1 - 23
Aligned: 23
Modelled: 23
Confidence: 6.0%
Identity: 30%
PDB header:transcription
Chain: B: PDB Molecule:hepatocyte nuclear factor 1-alpha;
PDBTitle: hnf-1alpha dimerization domain, with selenomethionine2 substitued at leu 12
Phyre2
| 15 |
|
PDB 1g2y chain A
Region: 1 - 23
Aligned: 23
Modelled: 23
Confidence: 5.9%
Identity: 30%
Fold: Dimerisation interlock
Superfamily: Dimerization cofactor of HNF-1 alpha
Family: Dimerization cofactor of HNF-1 alpha
Phyre2
| 16 |
|
PDB 1g2y chain A
Region: 1 - 23
Aligned: 23
Modelled: 23
Confidence: 5.9%
Identity: 30%
PDB header:transcription
Chain: A: PDB Molecule:hepatocyte nuclear factor 1-alpha;
PDBTitle: hnf-1alpha dimerization domain, with selenomethionine2 substitued at leu 12
Phyre2
| 17 |
|
PDB 7cei chain B
Region: 39 - 70
Aligned: 30
Modelled: 32
Confidence: 5.7%
Identity: 27%
PDB header:immune system
Chain: B: PDB Molecule:protein (colicin e7 immunity protein);
PDBTitle: the endonuclease domain of colicin e7 in complex with its inhibitor2 im7 protein
Phyre2