| 1 |
|
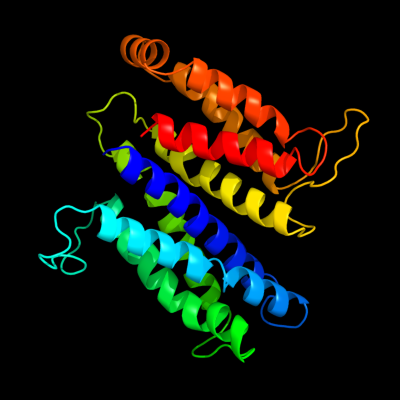
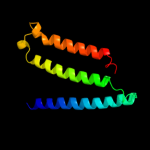
PDB 2vpz chain G
Region: 6 - 273
Aligned: 227
Modelled: 241
Confidence: 100.0%
Identity: 19%
PDB header:oxidoreductase
Chain: G: PDB Molecule:hypothetical membrane spanning protein;
PDBTitle: polysulfide reductase native structure
Phyre2
| 2 |
|
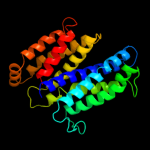
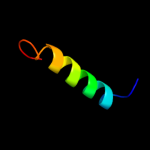
PDB 2jag chain A
Region: 147 - 279
Aligned: 133
Modelled: 133
Confidence: 56.0%
Identity: 12%
PDB header:membrane protein
Chain: A: PDB Molecule:halorhodopsin;
PDBTitle: l1-intermediate of halorhodopsin t203v
Phyre2
| 3 |
|
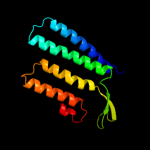
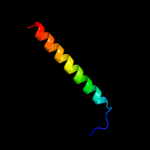
PDB 1e12 chain A
Region: 147 - 279
Aligned: 133
Modelled: 133
Confidence: 38.8%
Identity: 12%
Fold: Family A G protein-coupled receptor-like
Superfamily: Family A G protein-coupled receptor-like
Family: Bacteriorhodopsin-like
Phyre2
| 4 |
|
PDB 2amn chain A
Region: 8 - 18
Aligned: 11
Modelled: 11
Confidence: 13.6%
Identity: 55%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:cathelicidin;
PDBTitle: solution structure of fowlicidin-1, a novel cathelicidin2 antimicrobial peptide from chicken
Phyre2
| 5 |
|
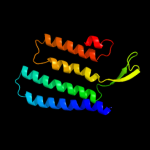
PDB 3rko chain A
Region: 13 - 113
Aligned: 93
Modelled: 101
Confidence: 11.1%
Identity: 13%
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh-quinone oxidoreductase subunit a;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 6 |
|
PDB 2p9x chain B
Region: 264 - 287
Aligned: 24
Modelled: 24
Confidence: 6.6%
Identity: 17%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ph0832;
PDBTitle: crystal structure of ph0832 from pyrococcus horikoshii ot3
Phyre2
| 7 |
|
PDB 2jwa chain A
Region: 109 - 143
Aligned: 35
Modelled: 35
Confidence: 6.4%
Identity: 17%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 8 |
|
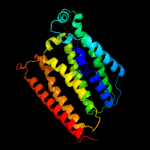
PDB 1m56 chain G
Region: 5 - 254
Aligned: 238
Modelled: 250
Confidence: 6.2%
Identity: 8%
PDB header:oxidoreductase
Chain: G: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure of cytochrome c oxidase from rhodobactor2 sphaeroides (wild type)
Phyre2