| 1 |
|
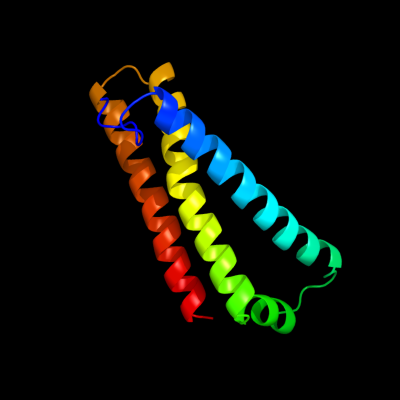
PDB 1nek chain D
Region: 3 - 115
Aligned: 113
Modelled: 113
Confidence: 100.0%
Identity: 100%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 2 |
|
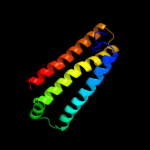
PDB 2bs2 chain C domain 1
Region: 3 - 114
Aligned: 112
Modelled: 112
Confidence: 99.3%
Identity: 11%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Fumarate reductase respiratory complex cytochrome b subunit, FrdC
Phyre2
| 3 |
|
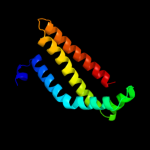
PDB 1nek chain C
Region: 12 - 111
Aligned: 100
Modelled: 100
Confidence: 99.0%
Identity: 17%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 4 |
|
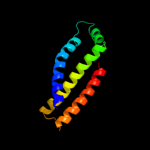
PDB 1zp0 chain D
Region: 8 - 111
Aligned: 85
Modelled: 88
Confidence: 99.0%
Identity: 14%
PDB header:oxidoreductase
Chain: D: PDB Molecule:small cytochrome binding protein;
PDBTitle: crystal structure of mitochondrial respiratory complex ii2 bound with 3-nitropropionate and 2-thenoyltrifluoroacetone
Phyre2
| 5 |
|
PDB 1yq3 chain C
Region: 13 - 107
Aligned: 95
Modelled: 95
Confidence: 98.5%
Identity: 16%
PDB header:oxidoreductase
Chain: C: PDB Molecule:succinate dehydrogenase cytochrome b, large subunit;
PDBTitle: avian respiratory complex ii with oxaloacetate and ubiquinone
Phyre2
| 6 |
|
PDB 1kf6 chain C
Region: 14 - 72
Aligned: 59
Modelled: 59
Confidence: 96.4%
Identity: 17%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 7 |
|
PDB 1kf6 chain D
Region: 21 - 107
Aligned: 87
Modelled: 87
Confidence: 95.6%
Identity: 20%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 8 |
|
PDB 1kcf chain A domain 1
Region: 4 - 21
Aligned: 18
Modelled: 18
Confidence: 10.6%
Identity: 33%
Fold: LEM/SAP HeH motif
Superfamily: SAP domain
Family: SAP domain
Phyre2
| 9 |
|
PDB 2do5 chain A
Region: 7 - 23
Aligned: 17
Modelled: 17
Confidence: 10.4%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:splicing factor 3b subunit 2;
PDBTitle: solution structure of the sap domain of human splicing2 factor 3b subunit 2
Phyre2
| 10 |
|
PDB 3da4 chain B
Region: 80 - 106
Aligned: 27
Modelled: 27
Confidence: 10.2%
Identity: 30%
PDB header:antibiotic
Chain: B: PDB Molecule:colicin-m;
PDBTitle: crystal structure of colicin m, a novel phosphatase2 specifically imported by escherichia coli
Phyre2
| 11 |
|
PDB 2gzd chain C
Region: 80 - 92
Aligned: 13
Modelled: 12
Confidence: 8.1%
Identity: 38%
PDB header:protein transport
Chain: C: PDB Molecule:rab11 family-interacting protein 2;
PDBTitle: crystal structure of rab11 in complex with rab11-fip2
Phyre2
| 12 |
|
PDB 1y8o chain A domain 1
Region: 65 - 84
Aligned: 20
Modelled: 20
Confidence: 6.8%
Identity: 5%
Fold: Bromodomain-like
Superfamily: alpha-ketoacid dehydrogenase kinase, N-terminal domain
Family: alpha-ketoacid dehydrogenase kinase, N-terminal domain
Phyre2
| 13 |
|
PDB 3mvn chain A
Region: 79 - 96
Aligned: 18
Modelled: 18
Confidence: 6.3%
Identity: 22%
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamayl-medo-
PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamayl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
Phyre2
| 14 |
|
PDB 2eyq chain A domain 6
Region: 81 - 94
Aligned: 14
Modelled: 14
Confidence: 5.4%
Identity: 21%
Fold: TRCF domain-like
Superfamily: TRCF domain-like
Family: TRCF domain
Phyre2