| Secondary structure and disorder prediction | |
| | |
1 | . | . | . | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | 20 | . | . | . | . | . | . | . | . | . | 30 | . | . | . | . | . | . | . | . | . | 40 | . | . | . | . | . | . | . | . | . | 50 | . | . | . | . | . | . | . | . | . | 60 |
| Sequence | |
M | N | I | L | L | S | I | A | I | T | T | G | I | L | S | G | I | W | G | W | V | A | V | S | L | G | L | L | S | W | A | G | F | L | G | C | T | A | Y | F | A | C | P | Q | G | G | L | K | G | L | A | I | S | A | A | T | L | L | S | G |
| Secondary structure | |
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 70 | . | . | . | . | . | . | . | . | . | 80 | . | . | . | . | . | . | . | . | . | 90 | . | . | . | . | . | . | . | . | . | 100 | . | . | . | . | . | . | . | . | . | 110 | . | . | . | . | . | . | . | . | . | 120 |
| Sequence | |
V | V | W | A | M | V | I | I | Y | G | S | A | L | A | P | H | L | E | I | L | G | Y | V | I | T | G | I | V | A | F | L | M | C | I | Q | A | K | Q | L | L | L | S | F | V | P | G | T | F | I | G | A | C | A | T | F | A | G | Q | G | D |
| Secondary structure | |
 |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
|
|
|
|
|
|
| ? | ? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 130 | . | . | . | . | . | . | . | . | . | 140 | . | . | . | . | . | . | . | . | . | 150 | . | . | . | . | . | . | . | . | . | 160 | . | . | . |
| Sequence | |
W | K | L | V | L | P | S | L | A | L | G | L | I | F | G | Y | A | M | K | N | S | G | L | W | L | A | A | R | S | A | K | T | A | H | R | E | Q | E | I | K | N | K | A |
| Secondary structure | |
 |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|  |  |  |  |  |  |  |  |  |
|
|
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? | ? | ? | ? | ? |
|
|
|
|
| ? | ? | ? |
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| Confidence Key |
| High(9) | |
|
|
|
|
|
|
|
|
|
Low (0) |
| ? | Disordered |
  | Alpha helix |
  | Beta strand |
Hover over an aligned region to see model and summary info
Please note, only up to the top 20 hits are modelled to reduce computer load
|
| 1 |
|
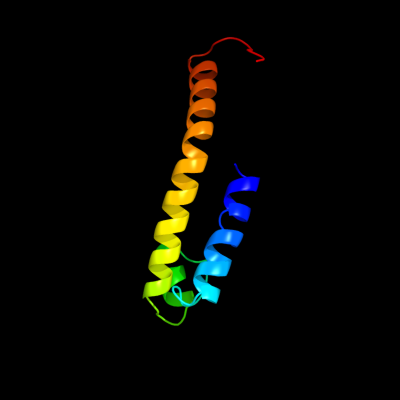
PDB 3qnq chain D
Region: 78 - 161
Aligned: 84
Modelled: 84
Confidence: 43.5%
Identity: 8%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 2 |
|
PDB 2wvm chain A
Region: 102 - 118
Aligned: 17
Modelled: 17
Confidence: 14.4%
Identity: 18%
PDB header:transferase
Chain: A: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: h309a mutant of mannosyl-3-phosphoglycerate synthase from2 thermus thermophilus hb27 in complex with3 gdp-alpha-d-mannose and mg(ii)
Phyre2
| 3 |
|
PDB 2zu8 chain A
Region: 102 - 118
Aligned: 17
Modelled: 17
Confidence: 13.1%
Identity: 18%
PDB header:transferase
Chain: A: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: crystal structure of mannosyl-3-phosphoglycerate synthase2 from pyrococcus horikoshii
Phyre2
| 4 |
|
PDB 2nww chain A domain 1
Region: 100 - 161
Aligned: 59
Modelled: 62
Confidence: 9.1%
Identity: 15%
Fold: Proton glutamate symport protein
Superfamily: Proton glutamate symport protein
Family: Proton glutamate symport protein
Phyre2
|
| Detailed template information | |
Due to computational demand, binding site predictions are not run for batch jobs
If you want to predict binding sites, please manually submit your model of choice to 3DLigandSite
Phyre is for academic use only
| Please cite: Protein structure prediction on
the web: a case study using the Phyre server |
| Kelley LA and Sternberg MJE. Nature Protocols
4, 363 - 371 (2009) [pdf] [Import into BibTeX] |
| |
| If you use the binding site
predictions from 3DLigandSite, please also cite: |
| 3DLigandSite: predicting ligand-binding sites using similar structures. |
| Wass MN, Kelley LA and Sternberg
MJ Nucleic Acids Research 38, W469-73 (2010) [PubMed] |
| |
|
|
|
|