| 1 |
|
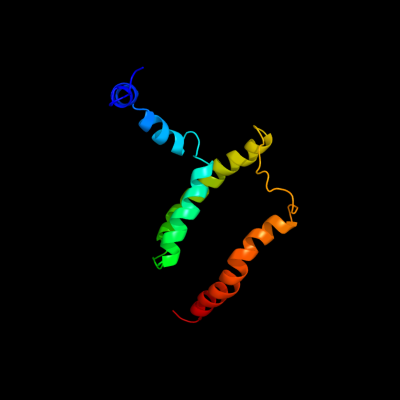
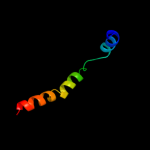
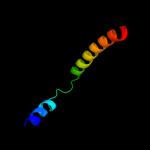
PDB 2kdc chain C
Region: 2 - 122
Aligned: 121
Modelled: 121
Confidence: 100.0%
Identity: 100%
PDB header:transferase
Chain: C: PDB Molecule:diacylglycerol kinase;
PDBTitle: nmr solution structure of e. coli diacylglycerol kinase2 (dagk) in dpc micelles
Phyre2
| 2 |
|
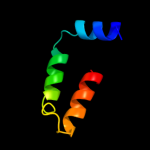
PDB 1rhz chain B
Region: 70 - 120
Aligned: 51
Modelled: 51
Confidence: 15.7%
Identity: 16%
Fold: Single transmembrane helix
Superfamily: Preprotein translocase SecE subunit
Family: Preprotein translocase SecE subunit
Phyre2
| 3 |
|
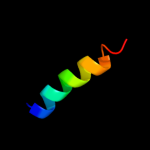
PDB 1ik7 chain A
Region: 60 - 91
Aligned: 32
Modelled: 32
Confidence: 13.5%
Identity: 13%
Fold: DEATH domain
Superfamily: DEATH domain
Family: DEATH domain, DD
Phyre2
| 4 |
|
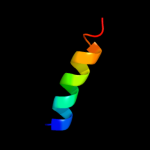
PDB 2y69 chain X
Region: 84 - 112
Aligned: 28
Modelled: 29
Confidence: 12.6%
Identity: 18%
PDB header:electron transport
Chain: X: PDB Molecule:cytochrome c oxidase polypeptide 7b;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 5 |
|
PDB 1v54 chain K
Region: 84 - 112
Aligned: 28
Modelled: 29
Confidence: 12.6%
Identity: 18%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIb
Family: Mitochondrial cytochrome c oxidase subunit VIIb
Phyre2
| 6 |
|
PDB 1rh5 chain B
Region: 70 - 119
Aligned: 50
Modelled: 50
Confidence: 7.0%
Identity: 16%
Fold: Single transmembrane helix
Superfamily: Preprotein translocase SecE subunit
Family: Preprotein translocase SecE subunit
Phyre2
| 7 |
|
PDB 2wwb chain B
Region: 70 - 111
Aligned: 42
Modelled: 42
Confidence: 6.9%
Identity: 12%
PDB header:ribosome
Chain: B: PDB Molecule:protein transport protein sec61 subunit gamma;
PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
Phyre2
| 8 |
|
PDB 1p58 chain F
Region: 23 - 66
Aligned: 44
Modelled: 44
Confidence: 6.4%
Identity: 16%
PDB header:virus
Chain: F: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
Phyre2
| 9 |
|
PDB 3kdp chain H
Region: 103 - 120
Aligned: 18
Modelled: 18
Confidence: 6.1%
Identity: 39%
PDB header:hydrolase
Chain: H: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 10 |
|
PDB 3kdp chain G
Region: 103 - 120
Aligned: 18
Modelled: 18
Confidence: 6.1%
Identity: 39%
PDB header:hydrolase
Chain: G: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 11 |
|
PDB 2ww9 chain B
Region: 70 - 111
Aligned: 42
Modelled: 42
Confidence: 6.0%
Identity: 17%
PDB header:ribosome
Chain: B: PDB Molecule:protein transport protein sss1;
PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the2 yeast 80s ribosome
Phyre2