| 1 |
|
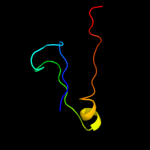
PDB 1edy chain A
Region: 1 - 24
Aligned: 24
Modelled: 24
Confidence: 28.0%
Identity: 29%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Alpha-macroglobulin receptor domain
Family: Alpha-macroglobulin receptor domain
Phyre2
| 2 |
|
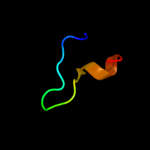
PDB 1bv8 chain A
Region: 1 - 24
Aligned: 24
Modelled: 24
Confidence: 27.4%
Identity: 29%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Alpha-macroglobulin receptor domain
Family: Alpha-macroglobulin receptor domain
Phyre2
| 3 |
|
PDB 1ayo chain A
Region: 1 - 24
Aligned: 24
Modelled: 24
Confidence: 24.1%
Identity: 33%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Alpha-macroglobulin receptor domain
Family: Alpha-macroglobulin receptor domain
Phyre2
| 4 |
|
PDB 1qhd chain A domain 2
Region: 118 - 142
Aligned: 25
Modelled: 24
Confidence: 19.6%
Identity: 48%
Fold: Viral protein domain
Superfamily: Viral protein domain
Family: Top domain of virus capsid protein
Phyre2
| 5 |
|
PDB 1qgc chain 1
Region: 30 - 58
Aligned: 29
Modelled: 29
Confidence: 19.2%
Identity: 28%
PDB header:virus/immune system
Chain: 1: PDB Molecule:protein (virus capsid protein);
PDB Fragment:residues 133-156
PDBTitle: structure of the complex of an fab fragment of a neutralizing antibody2 with foot and mouth disease virus
Phyre2
| 6 |
|
PDB 2knn chain A
Region: 76 - 98
Aligned: 22
Modelled: 23
Confidence: 12.7%
Identity: 45%
PDB header:plant protein
Chain: A: PDB Molecule:cycloviolacin-o2;
PDBTitle: solution structure of the cyclotide cycloviolacin o2 with2 glu6 methylated (cyo2me)
Phyre2
| 7 |
|
PDB 2wzr chain 1
Region: 32 - 73
Aligned: 36
Modelled: 41
Confidence: 12.6%
Identity: 39%
PDB header:virus
Chain: 1: PDB Molecule:polyprotein;
PDBTitle: the structure of foot and mouth disease virus serotype sat1
Phyre2
| 8 |
|
PDB 1dt9 chain A domain 3
Region: 10 - 27
Aligned: 17
Modelled: 18
Confidence: 10.9%
Identity: 47%
Fold: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Superfamily: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Family: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1
Phyre2
| 9 |
|
PDB 2jr7 chain A
Region: 69 - 78
Aligned: 8
Modelled: 10
Confidence: 10.2%
Identity: 63%
PDB header:metal binding protein
Chain: A: PDB Molecule:dph3 homolog;
PDBTitle: solution structure of human desr1
Phyre2
| 10 |
|
PDB 2jra chain B
Region: 59 - 71
Aligned: 13
Modelled: 13
Confidence: 7.9%
Identity: 46%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein rpa2121;
PDBTitle: a novel domain-swapped solution nmr structure of protein rpa2121 from2 rhodopseudomonas palustris. northeast structural genomics target rpt6
Phyre2
| 11 |
|
PDB 2kcg chain A
Region: 76 - 93
Aligned: 18
Modelled: 18
Confidence: 6.8%
Identity: 50%
PDB header:plant protein
Chain: A: PDB Molecule:cycloviolacin-o2;
PDBTitle: solution structure of cycloviolacin o2
Phyre2
| 12 |
|
PDB 2fug chain 7 domain 1
Region: 4 - 27
Aligned: 24
Modelled: 24
Confidence: 6.7%
Identity: 38%
Fold: N domain of copper amine oxidase-like
Superfamily: Frataxin/Nqo15-like
Family: Nqo15-like
Phyre2