| 1 |
|
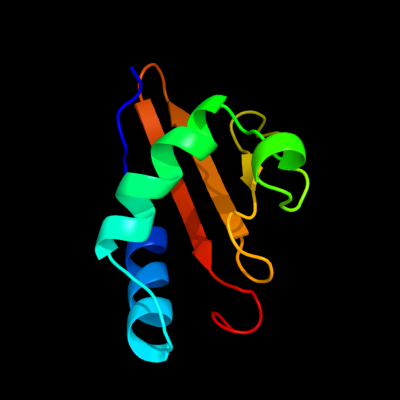
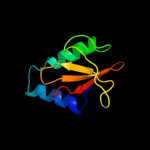
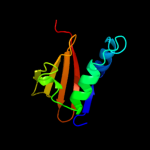
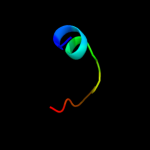
PDB 1z8m chain A domain 1
Region: 3 - 91
Aligned: 88
Modelled: 89
Confidence: 100.0%
Identity: 30%
Fold: RelE-like
Superfamily: RelE-like
Family: RelE-like
Phyre2
| 2 |
|
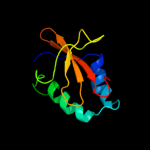
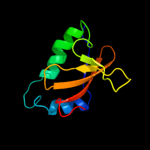
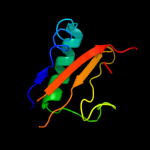
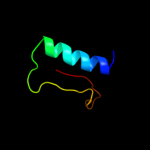
PDB 2otr chain A
Region: 3 - 91
Aligned: 88
Modelled: 89
Confidence: 100.0%
Identity: 36%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein hp0892;
PDBTitle: solution structure of conserved hypothetical protein hp0892 from2 helicobacter pylori
Phyre2
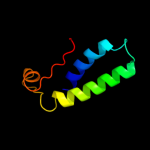
| 3 |
|
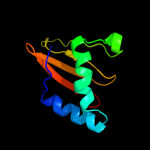
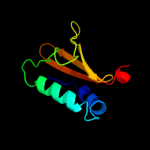
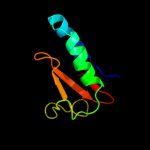
PDB 3oei chain H
Region: 3 - 87
Aligned: 83
Modelled: 85
Confidence: 99.9%
Identity: 18%
PDB header:toxin, protein binding
Chain: H: PDB Molecule:relk (toxin rv3358);
PDBTitle: crystal structure of mycobacterium tuberculosis reljk (rv3357-rv3358-2 relbe3)
Phyre2
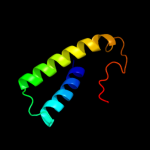
| 4 |
|
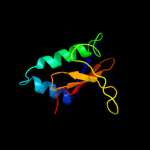
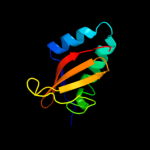
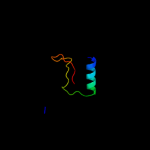
PDB 2a6s chain A domain 1
Region: 5 - 87
Aligned: 81
Modelled: 83
Confidence: 99.9%
Identity: 22%
Fold: RelE-like
Superfamily: RelE-like
Family: YoeB/Txe-like
Phyre2
| 5 |
|
PDB 2khe chain A
Region: 2 - 91
Aligned: 85
Modelled: 90
Confidence: 99.2%
Identity: 16%
PDB header:hydrolase
Chain: A: PDB Molecule:toxin-like protein;
PDBTitle: solution structure of the bacterial toxin rele from thermus2 thermophilus hb8
Phyre2
| 6 |
|
PDB 3bpq chain D
Region: 6 - 91
Aligned: 80
Modelled: 86
Confidence: 99.1%
Identity: 14%
PDB header:toxin
Chain: D: PDB Molecule:toxin rele3;
PDBTitle: crystal structure of relb-rele antitoxin-toxin complex from2 methanococcus jannaschii
Phyre2
| 7 |
|
PDB 3g5o chain C
Region: 1 - 89
Aligned: 82
Modelled: 89
Confidence: 98.7%
Identity: 17%
PDB header:toxin/antitoxin
Chain: C: PDB Molecule:uncharacterized protein rv2866;
PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
Phyre2
| 8 |
|
PDB 1wmi chain A domain 1
Region: 2 - 89
Aligned: 83
Modelled: 87
Confidence: 98.6%
Identity: 18%
Fold: RelE-like
Superfamily: RelE-like
Family: RelE-like
Phyre2
| 9 |
|
PDB 3kix chain Y
Region: 3 - 91
Aligned: 82
Modelled: 82
Confidence: 97.1%
Identity: 12%
PDB header:ribosome
Chain: Y: PDB Molecule:
PDBTitle: structure of rele nuclease bound to the 70s ribosome2 (postcleavage state; part 3 of 4)
Phyre2
| 10 |
|
PDB 3kxe chain B
Region: 4 - 81
Aligned: 71
Modelled: 78
Confidence: 91.6%
Identity: 14%
PDB header:protein binding
Chain: B: PDB Molecule:toxin protein pare-1;
PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
Phyre2
| 11 |
|
PDB 1vj7 chain A domain 2
Region: 24 - 64
Aligned: 41
Modelled: 41
Confidence: 32.4%
Identity: 17%
Fold: Nucleotidyltransferase
Superfamily: Nucleotidyltransferase
Family: RelA/SpoT domain
Phyre2
| 12 |
|
PDB 2rbd chain B
Region: 14 - 51
Aligned: 38
Modelled: 38
Confidence: 16.8%
Identity: 26%
PDB header:structural protein
Chain: B: PDB Molecule:bh2358 protein;
PDBTitle: crystal structure of a putative spore coat protein (bh2358) from2 bacillus halodurans c-125 at 1.54 a resolution
Phyre2
| 13 |
|
PDB 2y9m chain B
Region: 55 - 71
Aligned: 17
Modelled: 17
Confidence: 15.7%
Identity: 12%
PDB header:ligase/transport protein
Chain: B: PDB Molecule:peroxisome assembly protein 22;
PDBTitle: pex4p-pex22p structure
Phyre2
| 14 |
|
PDB 1vj7 chain B
Region: 24 - 64
Aligned: 41
Modelled: 41
Confidence: 15.4%
Identity: 17%
PDB header:hydrolase, transferase
Chain: B: PDB Molecule:bifunctional rela/spot;
PDBTitle: crystal structure of the bifunctional catalytic fragment of relseq,2 the rela/spot homolog from streptococcus equisimilis.
Phyre2
| 15 |
|
PDB 1jya chain A
Region: 58 - 73
Aligned: 15
Modelled: 12
Confidence: 15.0%
Identity: 27%
Fold: Secretion chaperone-like
Superfamily: Type III secretory system chaperone-like
Family: Type III secretory system chaperone
Phyre2
| 16 |
|
PDB 2z15 chain A domain 1
Region: 2 - 66
Aligned: 62
Modelled: 65
Confidence: 10.4%
Identity: 16%
Fold: BTG domain-like
Superfamily: BTG domain-like
Family: BTG domain-like
Phyre2
| 17 |
|
PDB 2d9g chain A
Region: 57 - 64
Aligned: 8
Modelled: 8
Confidence: 9.7%
Identity: 50%
PDB header:transcription
Chain: A: PDB Molecule:yy1-associated factor 2;
PDBTitle: solution structure of the zf-ranbp domain of yy1-associated2 factor 2
Phyre2
| 18 |
|
PDB 3e9v chain A domain 1
Region: 2 - 66
Aligned: 59
Modelled: 61
Confidence: 9.4%
Identity: 22%
Fold: BTG domain-like
Superfamily: BTG domain-like
Family: BTG domain-like
Phyre2
| 19 |
|
PDB 2ket chain A
Region: 10 - 22
Aligned: 13
Modelled: 13
Confidence: 6.6%
Identity: 23%
PDB header:antibiotic
Chain: A: PDB Molecule:cathelicidin-6;
PDBTitle: solution structure of bmap-27
Phyre2
| 20 |
|
PDB 2qn6 chain A domain 2
Region: 54 - 86
Aligned: 33
Modelled: 33
Confidence: 5.9%
Identity: 6%
Fold: Elongation factor/aminomethyltransferase common domain
Superfamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain
Family: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain
Phyre2