| 1 |
|
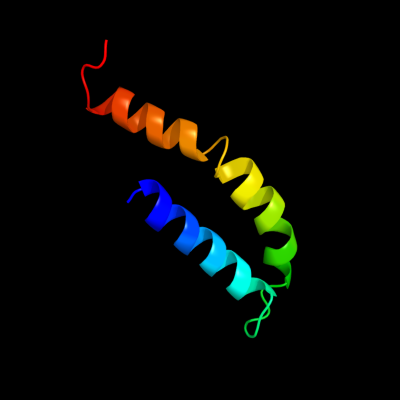
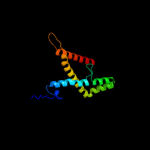
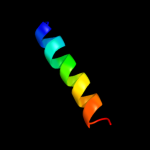
PDB 2r6g chain F domain 1
Region: 182 - 239
Aligned: 56
Modelled: 58
Confidence: 91.6%
Identity: 20%
Fold: MalF N-terminal region-like
Superfamily: MalF N-terminal region-like
Family: MalF N-terminal region-like
Phyre2
| 2 |
|
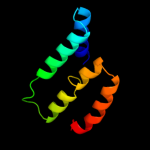
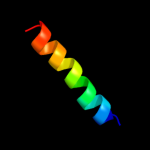
PDB 2xq2 chain A
Region: 29 - 290
Aligned: 222
Modelled: 241
Confidence: 23.2%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 3 |
|
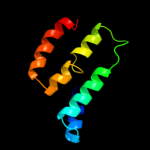
PDB 2qks chain A
Region: 172 - 262
Aligned: 90
Modelled: 91
Confidence: 11.3%
Identity: 17%
PDB header:metal transport
Chain: A: PDB Molecule:kir3.1-prokaryotic kir channel chimera;
PDBTitle: crystal structure of a kir3.1-prokaryotic kir channel chimera
Phyre2
| 4 |
|
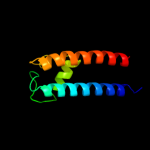
PDB 1kf6 chain C
Region: 4 - 158
Aligned: 126
Modelled: 136
Confidence: 7.0%
Identity: 13%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 5 |
|
PDB 1a87 chain A
Region: 17 - 92
Aligned: 74
Modelled: 76
Confidence: 6.6%
Identity: 18%
Fold: Toxins' membrane translocation domains
Superfamily: Colicin
Family: Colicin
Phyre2
| 6 |
|
PDB 1a87 chain A
Region: 17 - 92
Aligned: 74
Modelled: 76
Confidence: 6.6%
Identity: 18%
PDB header:bacteriocin
Chain: A: PDB Molecule:colicin n;
PDBTitle: colicin n
Phyre2
| 7 |
|
PDB 1xl6 chain B
Region: 172 - 262
Aligned: 90
Modelled: 91
Confidence: 6.3%
Identity: 11%
PDB header:metal transport
Chain: B: PDB Molecule:inward rectifier potassium channel;
PDBTitle: intermediate gating structure 2 of the inwardly rectifying k+ channel2 kirbac3.1
Phyre2
| 8 |
|
PDB 3b8e chain B
Region: 238 - 257
Aligned: 20
Modelled: 20
Confidence: 6.2%
Identity: 40%
PDB header:hydrolase/transport protein
Chain: B: PDB Molecule:sodium/potassium-transporting atpase subunit
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 9 |
|
PDB 3ixz chain B
Region: 238 - 258
Aligned: 21
Modelled: 21
Confidence: 5.4%
Identity: 29%
PDB header:hydrolase
Chain: B: PDB Molecule:potassium-transporting atpase subunit beta;
PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
Phyre2