| 1 |
|
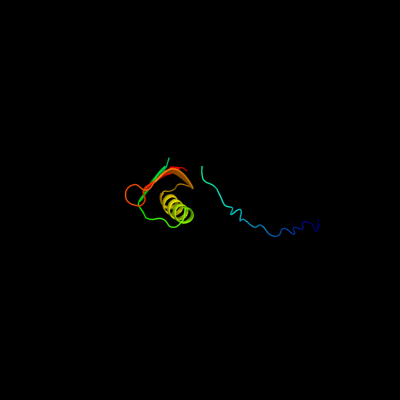
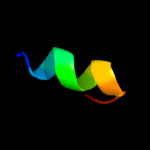
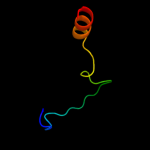
PDB 2jna chain A domain 1
Region: 3 - 78
Aligned: 76
Modelled: 76
Confidence: 97.8%
Identity: 30%
Fold: Dodecin subunit-like
Superfamily: YdgH-like
Family: YdgH-like
Phyre2
| 2 |
|
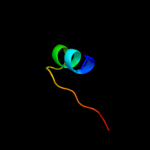
PDB 2noc chain A domain 1
Region: 28 - 78
Aligned: 50
Modelled: 51
Confidence: 95.9%
Identity: 26%
Fold: Dodecin subunit-like
Superfamily: YdgH-like
Family: YdgH-like
Phyre2
| 3 |
|
PDB 2l69 chain A
Region: 39 - 56
Aligned: 18
Modelled: 18
Confidence: 25.3%
Identity: 50%
PDB header:de novo protein
Chain: A: PDB Molecule:rossmann 2x3 fold protein;
PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or28
Phyre2
| 4 |
|
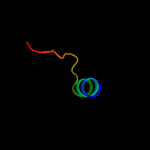
PDB 1y7m chain A domain 2
Region: 45 - 57
Aligned: 13
Modelled: 13
Confidence: 20.4%
Identity: 38%
Fold: LysM domain
Superfamily: LysM domain
Family: LysM domain
Phyre2
| 5 |
|
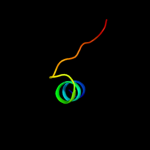
PDB 1e0g chain A
Region: 45 - 57
Aligned: 13
Modelled: 13
Confidence: 18.8%
Identity: 23%
Fold: LysM domain
Superfamily: LysM domain
Family: LysM domain
Phyre2
| 6 |
|
PDB 2djp chain A
Region: 45 - 57
Aligned: 13
Modelled: 13
Confidence: 15.2%
Identity: 46%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein sb145;
PDBTitle: the solution structure of the lysm domain of human2 hypothetical protein sb145
Phyre2
| 7 |
|
PDB 3c7b chain B domain 2
Region: 45 - 63
Aligned: 19
Modelled: 19
Confidence: 14.9%
Identity: 37%
Fold: Ferredoxin-like
Superfamily: Nitrite/Sulfite reductase N-terminal domain-like
Family: DsrA/DsrB N-terminal-domain-like
Phyre2
| 8 |
|
PDB 1zj8 chain A domain 1
Region: 45 - 63
Aligned: 19
Modelled: 19
Confidence: 10.6%
Identity: 16%
Fold: Ferredoxin-like
Superfamily: Nitrite/Sulfite reductase N-terminal domain-like
Family: Duplicated SiR/NiR-like domains 1 and 3
Phyre2
| 9 |
|
PDB 4a1d chain K
Region: 43 - 55
Aligned: 13
Modelled: 13
Confidence: 9.5%
Identity: 62%
PDB header:ribosome
Chain: K: PDB Molecule:ubiquitin-60s ribosomal protein l40;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
Phyre2
| 10 |
|
PDB 1xc0 chain A
Region: 5 - 15
Aligned: 11
Modelled: 11
Confidence: 8.6%
Identity: 73%
PDB header:signaling protein
Chain: A: PDB Molecule:pardaxin p-4;
PDBTitle: twenty lowest energy structures of pa4 by solution nmr
Phyre2
| 11 |
|
PDB 2v4j chain B domain 2
Region: 45 - 63
Aligned: 19
Modelled: 19
Confidence: 8.4%
Identity: 32%
Fold: Ferredoxin-like
Superfamily: Nitrite/Sulfite reductase N-terminal domain-like
Family: DsrA/DsrB N-terminal-domain-like
Phyre2
| 12 |
|
PDB 3sb1 chain B
Region: 21 - 53
Aligned: 33
Modelled: 33
Confidence: 8.0%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hydrogenase expression protein;
PDBTitle: hydrogenase expression protein huph from thiobacillus denitrificans2 atcc 25259
Phyre2