| 1 |
|
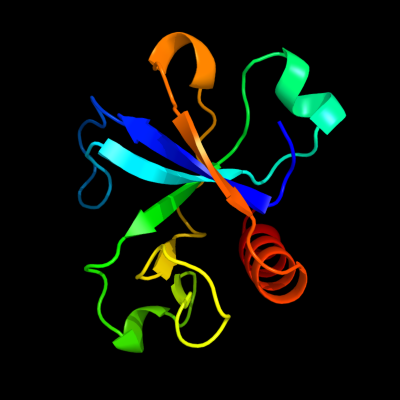
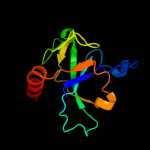
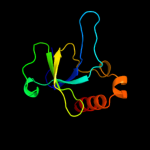
PDB 1m1f chain A
Region: 7 - 115
Aligned: 105
Modelled: 109
Confidence: 100.0%
Identity: 40%
Fold: SH3-like barrel
Superfamily: Cell growth inhibitor/plasmid maintenance toxic component
Family: Kid/PemK
Phyre2
| 2 |
|
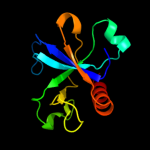
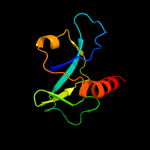
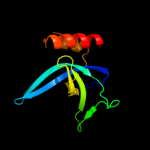
PDB 1ub4 chain A
Region: 2 - 115
Aligned: 101
Modelled: 114
Confidence: 100.0%
Identity: 31%
Fold: SH3-like barrel
Superfamily: Cell growth inhibitor/plasmid maintenance toxic component
Family: Kid/PemK
Phyre2
| 3 |
|
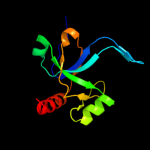
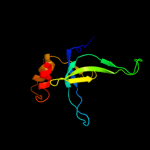
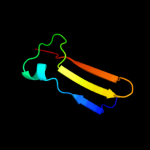
PDB 1ne8 chain A
Region: 5 - 116
Aligned: 110
Modelled: 112
Confidence: 100.0%
Identity: 25%
Fold: SH3-like barrel
Superfamily: Cell growth inhibitor/plasmid maintenance toxic component
Family: Kid/PemK
Phyre2
| 4 |
|
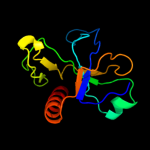
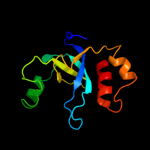
PDB 3jrz chain A
Region: 9 - 115
Aligned: 95
Modelled: 107
Confidence: 97.2%
Identity: 9%
PDB header:toxin
Chain: A: PDB Molecule:ccdb;
PDBTitle: ccdbvfi-formii-ph5.6
Phyre2
| 5 |
|
PDB 3vub chain A
Region: 26 - 116
Aligned: 89
Modelled: 91
Confidence: 96.7%
Identity: 7%
Fold: SH3-like barrel
Superfamily: Cell growth inhibitor/plasmid maintenance toxic component
Family: CcdB
Phyre2
| 6 |
|
PDB 2l89 chain A
Region: 2 - 116
Aligned: 95
Modelled: 104
Confidence: 83.5%
Identity: 21%
PDB header:protein binding
Chain: A: PDB Molecule:pwwp domain-containing protein 1;
PDBTitle: solution structure of pdp1 pwwp domain reveals its unique binding2 sites for methylated h4k20 and dna
Phyre2
| 7 |
|
PDB 1h3z chain A
Region: 2 - 116
Aligned: 97
Modelled: 115
Confidence: 83.1%
Identity: 9%
Fold: SH3-like barrel
Superfamily: Tudor/PWWP/MBT
Family: PWWP domain
Phyre2
| 8 |
|
PDB 3llr chain A
Region: 5 - 114
Aligned: 88
Modelled: 97
Confidence: 35.7%
Identity: 15%
PDB header:transferase
Chain: A: PDB Molecule:dna (cytosine-5)-methyltransferase 3a;
PDBTitle: crystal structure of the pwwp domain of human dna (cytosine-5-)-2 methyltransferase 3 alpha
Phyre2
| 9 |
|
PDB 2gfu chain A
Region: 3 - 114
Aligned: 92
Modelled: 100
Confidence: 23.1%
Identity: 20%
PDB header:dna binding protein
Chain: A: PDB Molecule:dna mismatch repair protein msh6;
PDBTitle: nmr solution structure of the pwwp domain of mismatch2 repair protein hmsh6
Phyre2
| 10 |
|
PDB 3mxu chain A
Region: 10 - 52
Aligned: 38
Modelled: 43
Confidence: 21.5%
Identity: 26%
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycine cleavage system h protein;
PDBTitle: crystal structure of glycine cleavage system protein h from bartonella2 henselae
Phyre2
| 11 |
|
PDB 2jys chain A
Region: 8 - 17
Aligned: 10
Modelled: 10
Confidence: 12.8%
Identity: 20%
PDB header:hydrolase
Chain: A: PDB Molecule:protease/reverse transcriptase;
PDBTitle: solution structure of simian foamy virus (mac) protease
Phyre2
| 12 |
|
PDB 1zak chain A domain 2
Region: 4 - 28
Aligned: 25
Modelled: 25
Confidence: 12.0%
Identity: 16%
Fold: Rubredoxin-like
Superfamily: Microbial and mitochondrial ADK, insert "zinc finger" domain
Family: Microbial and mitochondrial ADK, insert "zinc finger" domain
Phyre2
| 13 |
|
PDB 2nlu chain A domain 1
Region: 2 - 110
Aligned: 85
Modelled: 95
Confidence: 10.9%
Identity: 11%
Fold: SH3-like barrel
Superfamily: Tudor/PWWP/MBT
Family: PWWP domain
Phyre2
| 14 |
|
PDB 2jns chain A
Region: 85 - 112
Aligned: 28
Modelled: 28
Confidence: 10.5%
Identity: 14%
PDB header:unknown function
Chain: A: PDB Molecule:bromodomain-containing protein 4;
PDBTitle: solution structure of the bromodomain-containing protein 42 et domain
Phyre2
| 15 |
|
PDB 1hpc chain A
Region: 10 - 39
Aligned: 29
Modelled: 30
Confidence: 8.6%
Identity: 31%
Fold: Barrel-sandwich hybrid
Superfamily: Single hybrid motif
Family: Biotinyl/lipoyl-carrier proteins and domains
Phyre2
| 16 |
|
PDB 2eaa chain B
Region: 6 - 36
Aligned: 31
Modelled: 31
Confidence: 7.4%
Identity: 6%
PDB header:plant protein
Chain: B: PDB Molecule:7s globulin-3;
PDBTitle: crystal structure of adzuki bean 7s globulin-3
Phyre2
| 17 |
|
PDB 2daq chain A domain 1
Region: 6 - 114
Aligned: 90
Modelled: 109
Confidence: 7.2%
Identity: 20%
Fold: SH3-like barrel
Superfamily: Tudor/PWWP/MBT
Family: PWWP domain
Phyre2
| 18 |
|
PDB 1kp0 chain A domain 2
Region: 3 - 34
Aligned: 32
Modelled: 32
Confidence: 6.5%
Identity: 19%
Fold: Creatinase/aminopeptidase
Superfamily: Creatinase/aminopeptidase
Family: Creatinase/aminopeptidase
Phyre2
| 19 |
|
PDB 1zzo chain A domain 1
Region: 86 - 111
Aligned: 26
Modelled: 26
Confidence: 6.5%
Identity: 15%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione peroxidase-like
Phyre2
| 20 |
|
PDB 1b12 chain A
Region: 5 - 22
Aligned: 18
Modelled: 18
Confidence: 6.1%
Identity: 28%
Fold: LexA/Signal peptidase
Superfamily: LexA/Signal peptidase
Family: Type 1 signal peptidase
Phyre2