| 1 |
|
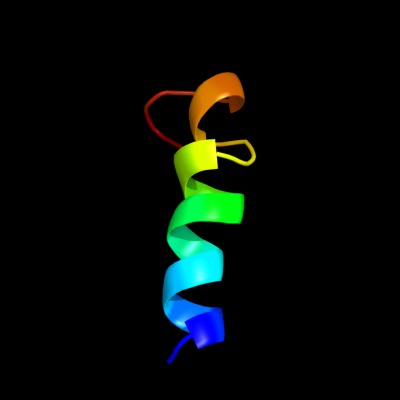
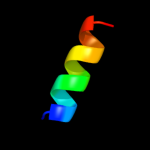
PDB 3fka chain D
Region: 2 - 25
Aligned: 23
Modelled: 24
Confidence: 24.9%
Identity: 9%
PDB header:unknown function
Chain: D: PDB Molecule:uncharacterized ntf-2 like protein;
PDBTitle: crystal structure of a ntf-2 like protein of unknown function2 (spo1084) from silicibacter pomeroyi dss-3 at 1.69 a resolution
Phyre2
| 2 |
|
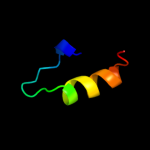
PDB 3duk chain D
Region: 2 - 25
Aligned: 23
Modelled: 24
Confidence: 24.0%
Identity: 26%
PDB header:unknown function
Chain: D: PDB Molecule:ntf2-like protein of unknown function;
PDBTitle: crystal structure of a ntf2-like protein of unknown function2 (mfla_0564) from methylobacillus flagellatus kt at 2.200 a resolution
Phyre2
| 3 |
|
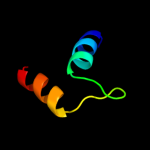
PDB 1k94 chain A
Region: 13 - 36
Aligned: 24
Modelled: 24
Confidence: 21.8%
Identity: 21%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Penta-EF-hand proteins
Phyre2
| 4 |
|
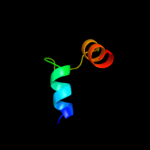
PDB 2qti chain A domain 1
Region: 28 - 41
Aligned: 14
Modelled: 14
Confidence: 18.3%
Identity: 50%
Fold: YejL-like
Superfamily: YejL-like
Family: YejL-like
Phyre2
| 5 |
|
PDB 2juw chain A domain 1
Region: 28 - 41
Aligned: 14
Modelled: 14
Confidence: 15.8%
Identity: 50%
Fold: YejL-like
Superfamily: YejL-like
Family: YejL-like
Phyre2
| 6 |
|
PDB 2jxc chain A domain 1
Region: 8 - 37
Aligned: 30
Modelled: 30
Confidence: 15.6%
Identity: 27%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Eps15 homology domain (EH domain)
Phyre2
| 7 |
|
PDB 2kq6 chain A
Region: 16 - 40
Aligned: 25
Modelled: 25
Confidence: 13.5%
Identity: 32%
PDB header:transport protein
Chain: A: PDB Molecule:polycystin-2;
PDBTitle: the structure of the ef-hand domain of polycystin-2 suggests a2 mechanism for ca2+-dependent regulation of polycystin-2 channel3 activity
Phyre2
| 8 |
|
PDB 2kfh chain A
Region: 8 - 37
Aligned: 30
Modelled: 30
Confidence: 11.3%
Identity: 20%
PDB header:protein binding
Chain: A: PDB Molecule:eh domain-containing protein 1;
PDBTitle: structure of the c-terminal domain of ehd1 with fnyestgpftak
Phyre2
| 9 |
|
PDB 1dvi chain A
Region: 8 - 36
Aligned: 29
Modelled: 29
Confidence: 11.3%
Identity: 24%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Penta-EF-hand proteins
Phyre2
| 10 |
|
PDB 1alv chain A
Region: 8 - 36
Aligned: 29
Modelled: 29
Confidence: 10.0%
Identity: 21%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Penta-EF-hand proteins
Phyre2
| 11 |
|
PDB 1mqs chain B
Region: 28 - 41
Aligned: 14
Modelled: 14
Confidence: 8.8%
Identity: 50%
PDB header:endocytosis/exocytosis
Chain: B: PDB Molecule:integral membrane protein sed5;
PDBTitle: crystal structure of sly1p in complex with an n-terminal2 peptide of sed5p
Phyre2
| 12 |
|
PDB 1g33 chain A
Region: 20 - 37
Aligned: 18
Modelled: 18
Confidence: 8.3%
Identity: 22%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Parvalbumin
Phyre2
| 13 |
|
PDB 1gjy chain A
Region: 13 - 36
Aligned: 24
Modelled: 24
Confidence: 8.2%
Identity: 21%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Penta-EF-hand proteins
Phyre2
| 14 |
|
PDB 3fhk chain F
Region: 7 - 20
Aligned: 14
Modelled: 14
Confidence: 8.0%
Identity: 50%
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:upf0403 protein yphp;
PDBTitle: crystal structure of apc1446, b.subtilis yphp disulfide2 isomerase
Phyre2
| 15 |
|
PDB 1qjt chain A
Region: 11 - 37
Aligned: 27
Modelled: 27
Confidence: 7.4%
Identity: 22%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Eps15 homology domain (EH domain)
Phyre2
| 16 |
|
PDB 2kle chain A
Region: 16 - 36
Aligned: 21
Modelled: 21
Confidence: 7.2%
Identity: 38%
PDB header:membrane protein
Chain: A: PDB Molecule:polycystin-2;
PDBTitle: isic refined solution structure of the calcium binding2 domain of the c-terminal cytosolic domain of polycystin-2
Phyre2
| 17 |
|
PDB 1kxp chain D domain 3
Region: 25 - 41
Aligned: 17
Modelled: 17
Confidence: 5.9%
Identity: 29%
Fold: Serum albumin-like
Superfamily: Serum albumin-like
Family: Serum albumin-like
Phyre2
| 18 |
|
PDB 1lkj chain A
Region: 8 - 35
Aligned: 28
Modelled: 28
Confidence: 5.6%
Identity: 14%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Calmodulin-like
Phyre2
| 19 |
|
PDB 2pmy chain B
Region: 8 - 37
Aligned: 30
Modelled: 30
Confidence: 5.5%
Identity: 30%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:ras and ef-hand domain-containing protein;
PDBTitle: ef-hand domain of human rasef
Phyre2
| 20 |
|
PDB 2vrg chain A
Region: 19 - 34
Aligned: 16
Modelled: 16
Confidence: 5.4%
Identity: 19%
PDB header:transport
Chain: A: PDB Molecule:multiple coagulation factor deficiency protein 2;
PDBTitle: structure of human mcfd2
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|
| 25 |
|