| 1 |
|
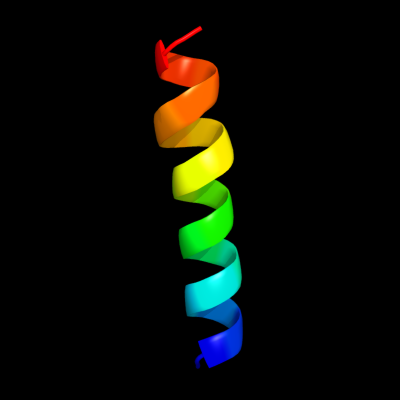
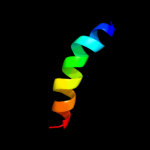
PDB 2zxe chain G
Region: 50 - 71
Aligned: 21
Modelled: 22
Confidence: 21.1%
Identity: 38%
PDB header:hydrolase/transport protein
Chain: G: PDB Molecule:phospholemman-like protein;
PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
Phyre2
| 2 |
|
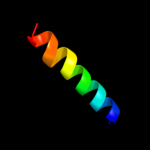
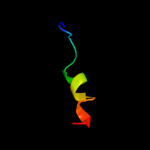
PDB 3n23 chain E
Region: 50 - 72
Aligned: 22
Modelled: 23
Confidence: 15.2%
Identity: 32%
PDB header:hydrolase
Chain: E: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the high affinity complex between ouabain and the2 e2p form of the sodium-potassium pump
Phyre2
| 3 |
|
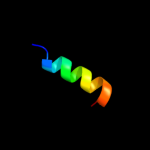
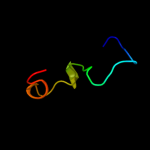
PDB 3vub chain A
Region: 55 - 71
Aligned: 17
Modelled: 17
Confidence: 12.8%
Identity: 24%
Fold: SH3-like barrel
Superfamily: Cell growth inhibitor/plasmid maintenance toxic component
Family: CcdB
Phyre2
| 4 |
|
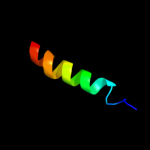
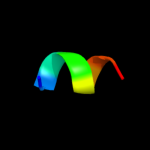
PDB 3jrz chain A
Region: 56 - 71
Aligned: 16
Modelled: 16
Confidence: 9.9%
Identity: 50%
PDB header:toxin
Chain: A: PDB Molecule:ccdb;
PDBTitle: ccdbvfi-formii-ph5.6
Phyre2
| 5 |
|
PDB 2jo1 chain A
Region: 50 - 72
Aligned: 22
Modelled: 23
Confidence: 9.0%
Identity: 45%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 6 |
|
PDB 2jp3 chain A
Region: 50 - 71
Aligned: 21
Modelled: 22
Confidence: 8.5%
Identity: 29%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 7 |
|
PDB 2l3y chain A
Region: 65 - 83
Aligned: 19
Modelled: 19
Confidence: 7.3%
Identity: 42%
PDB header:transcription
Chain: A: PDB Molecule:interleukin-6;
PDBTitle: solution structure of mouse il-6
Phyre2
| 8 |
|
PDB 2gv8 chain A domain 2
Region: 5 - 30
Aligned: 26
Modelled: 26
Confidence: 7.0%
Identity: 23%
Fold: FAD/NAD(P)-binding domain
Superfamily: FAD/NAD(P)-binding domain
Family: FAD/NAD-linked reductases, N-terminal and central domains
Phyre2
| 9 |
|
PDB 1ail chain A
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 6.1%
Identity: 63%
Fold: S15/NS1 RNA-binding domain
Superfamily: S15/NS1 RNA-binding domain
Family: N-terminal, RNA-binding domain of nonstructural protein NS1
Phyre2
| 10 |
|
PDB 3lfk chain C
Region: 33 - 54
Aligned: 22
Modelled: 22
Confidence: 5.8%
Identity: 50%
PDB header:unknown function
Chain: C: PDB Molecule:marr like protein, tvg0766549;
PDBTitle: a reported archaeal mechanosensitive channel is a structural2 homolog of marr-like transcriptional regulators
Phyre2