| 1 |
|
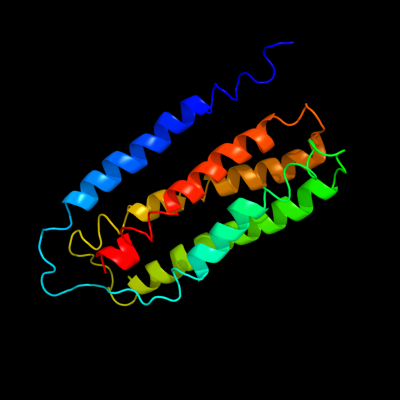
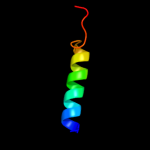
PDB 1fft chain C
Region: 19 - 203
Aligned: 185
Modelled: 185
Confidence: 100.0%
Identity: 100%
Fold: Cytochrome c oxidase subunit III-like
Superfamily: Cytochrome c oxidase subunit III-like
Family: Cytochrome c oxidase subunit III-like
Phyre2
| 2 |
|
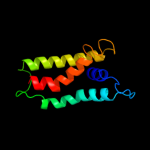
PDB 1v54 chain C
Region: 9 - 203
Aligned: 192
Modelled: 195
Confidence: 100.0%
Identity: 21%
Fold: Cytochrome c oxidase subunit III-like
Superfamily: Cytochrome c oxidase subunit III-like
Family: Cytochrome c oxidase subunit III-like
Phyre2
| 3 |
|
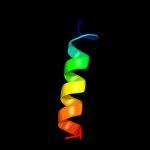
PDB 1qle chain C
Region: 15 - 202
Aligned: 185
Modelled: 188
Confidence: 100.0%
Identity: 25%
Fold: Cytochrome c oxidase subunit III-like
Superfamily: Cytochrome c oxidase subunit III-like
Family: Cytochrome c oxidase subunit III-like
Phyre2
| 4 |
|
PDB 1m56 chain C
Region: 15 - 202
Aligned: 185
Modelled: 188
Confidence: 100.0%
Identity: 28%
Fold: Cytochrome c oxidase subunit III-like
Superfamily: Cytochrome c oxidase subunit III-like
Family: Cytochrome c oxidase subunit III-like
Phyre2
| 5 |
|
PDB 2knc chain A
Region: 150 - 174
Aligned: 25
Modelled: 25
Confidence: 14.3%
Identity: 28%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 6 |
|
PDB 2k1a chain A
Region: 150 - 174
Aligned: 25
Modelled: 25
Confidence: 11.8%
Identity: 28%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
Phyre2
| 7 |
|
PDB 3cx5 chain C domain 1
Region: 43 - 147
Aligned: 97
Modelled: 105
Confidence: 7.2%
Identity: 15%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 8 |
|
PDB 1jb0 chain I
Region: 97 - 116
Aligned: 20
Modelled: 20
Confidence: 6.2%
Identity: 35%
Fold: Single transmembrane helix
Superfamily: Subunit VIII of photosystem I reaction centre, PsaI
Family: Subunit VIII of photosystem I reaction centre, PsaI
Phyre2