| 1 |
|
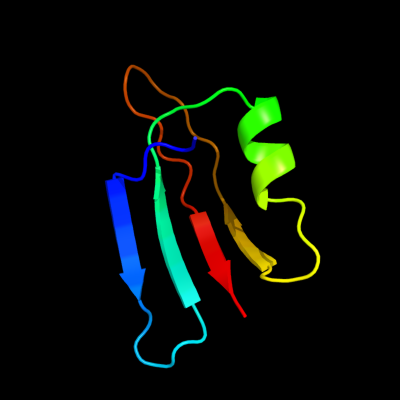
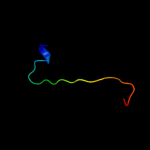
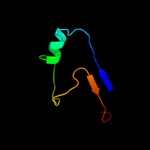
PDB 1uv7 chain A
Region: 84 - 142
Aligned: 56
Modelled: 59
Confidence: 89.6%
Identity: 20%
PDB header:transport
Chain: A: PDB Molecule:general secretion pathway protein m;
PDBTitle: periplasmic domain of epsm from vibrio cholerae
Phyre2
| 2 |
|
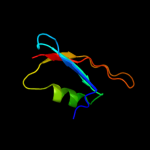
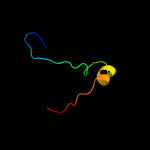
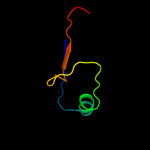
PDB 1uv7 chain A
Region: 84 - 142
Aligned: 56
Modelled: 59
Confidence: 89.6%
Identity: 20%
Fold: RRF/tRNA synthetase additional domain-like
Superfamily: General secretion pathway protein M, EpsM
Family: General secretion pathway protein M, EpsM
Phyre2
| 3 |
|
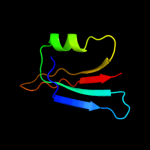
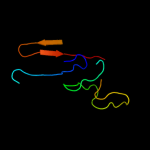
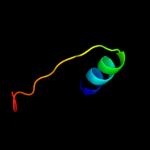
PDB 2rjz chain A
Region: 39 - 140
Aligned: 101
Modelled: 102
Confidence: 70.3%
Identity: 10%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:pilo protein;
PDBTitle: crystal structure of the type 4 fimbrial biogenesis protein pilo from2 pseudomonas aeruginosa
Phyre2
| 4 |
|
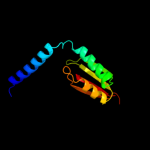
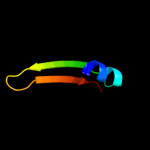
PDB 2e26 chain A
Region: 77 - 98
Aligned: 22
Modelled: 22
Confidence: 16.2%
Identity: 23%
PDB header:signaling protein
Chain: A: PDB Molecule:reelin;
PDBTitle: crystal structure of two repeat fragment of reelin
Phyre2
| 5 |
|
PDB 1ikp chain A domain 2
Region: 92 - 124
Aligned: 25
Modelled: 33
Confidence: 13.8%
Identity: 44%
Fold: ADP-ribosylation
Superfamily: ADP-ribosylation
Family: ADP-ribosylating toxins
Phyre2
| 6 |
|
PDB 2htb chain B
Region: 80 - 142
Aligned: 63
Modelled: 63
Confidence: 13.5%
Identity: 24%
PDB header:isomerase
Chain: B: PDB Molecule:putative enzyme related to aldose 1-epimerase;
PDBTitle: crystal structure of a putative mutarotase (yead) from2 salmonella typhimurium in monoclinic form
Phyre2
| 7 |
|
PDB 3sbt chain B
Region: 71 - 97
Aligned: 27
Modelled: 27
Confidence: 12.4%
Identity: 11%
PDB header:splicing
Chain: B: PDB Molecule:a1 cistron-splicing factor aar2;
PDBTitle: crystal structure of a aar2-prp8 complex
Phyre2
| 8 |
|
PDB 2et1 chain A domain 1
Region: 78 - 95
Aligned: 18
Modelled: 18
Confidence: 11.7%
Identity: 33%
Fold: Double-stranded beta-helix
Superfamily: RmlC-like cupins
Family: Germin/Seed storage 7S protein
Phyre2
| 9 |
|
PDB 2css chain A domain 1
Region: 88 - 99
Aligned: 12
Modelled: 12
Confidence: 9.6%
Identity: 50%
Fold: PDZ domain-like
Superfamily: PDZ domain-like
Family: PDZ domain
Phyre2
| 10 |
|
PDB 1fxz chain A domain 2
Region: 78 - 95
Aligned: 18
Modelled: 18
Confidence: 8.8%
Identity: 6%
Fold: Double-stranded beta-helix
Superfamily: RmlC-like cupins
Family: Germin/Seed storage 7S protein
Phyre2
| 11 |
|
PDB 1r89 chain A domain 3
Region: 110 - 142
Aligned: 33
Modelled: 33
Confidence: 8.6%
Identity: 12%
Fold: Ferredoxin-like
Superfamily: PAP/Archaeal CCA-adding enzyme, C-terminal domain
Family: Archaeal tRNA CCA-adding enzyme
Phyre2
| 12 |
|
PDB 1lc0 chain A domain 2
Region: 119 - 146
Aligned: 28
Modelled: 28
Confidence: 8.4%
Identity: 32%
Fold: FwdE/GAPDH domain-like
Superfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family: Biliverdin reductase
Phyre2
| 13 |
|
PDB 1od5 chain A domain 2
Region: 78 - 95
Aligned: 18
Modelled: 18
Confidence: 8.3%
Identity: 17%
Fold: Double-stranded beta-helix
Superfamily: RmlC-like cupins
Family: Germin/Seed storage 7S protein
Phyre2
| 14 |
|
PDB 1lsh chain B
Region: 99 - 144
Aligned: 46
Modelled: 46
Confidence: 8.0%
Identity: 20%
PDB header:lipid binding protein
Chain: B: PDB Molecule:lipovitellin (lv-2);
PDBTitle: lipid-protein interactions in lipovitellin
Phyre2
| 15 |
|
PDB 1lsh chain B
Region: 99 - 144
Aligned: 46
Modelled: 46
Confidence: 8.0%
Identity: 20%
Fold: Lipovitellin-phosvitin complex; beta-sheet shell regions
Superfamily: Lipovitellin-phosvitin complex; beta-sheet shell regions
Family: Lipovitellin-phosvitin complex; beta-sheet shell regions
Phyre2
| 16 |
|
PDB 2xte chain H
Region: 111 - 132
Aligned: 22
Modelled: 22
Confidence: 7.1%
Identity: 27%
PDB header:transcription
Chain: H: PDB Molecule:f-box-like/wd repeat-containing protein tbl1x;
PDBTitle: structure of the tbl1 tetramerisation domain
Phyre2
| 17 |
|
PDB 2xtd chain B
Region: 111 - 132
Aligned: 22
Modelled: 22
Confidence: 7.0%
Identity: 27%
PDB header:transcription
Chain: B: PDB Molecule:tbl1 f-box-like/wd repeat-containing protein tbl1x;
PDBTitle: structure of the tbl1 tetramerisation domain
Phyre2
| 18 |
|
PDB 2p5k chain A domain 1
Region: 111 - 130
Aligned: 20
Modelled: 20
Confidence: 5.9%
Identity: 15%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Arginine repressor (ArgR), N-terminal DNA-binding domain
Phyre2
| 19 |
|
PDB 2csp chain A domain 1
Region: 70 - 92
Aligned: 23
Modelled: 23
Confidence: 5.7%
Identity: 35%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Fibronectin type III
Family: Fibronectin type III
Phyre2
| 20 |
|
PDB 3ktb chain D
Region: 108 - 130
Aligned: 23
Modelled: 23
Confidence: 5.4%
Identity: 22%
PDB header:transcription regulator
Chain: D: PDB Molecule:arsenical resistance operon trans-acting repressor;
PDBTitle: crystal structure of arsenical resistance operon trans-acting2 repressor from bacteroides vulgatus atcc 8482
Phyre2