| 1 |
|
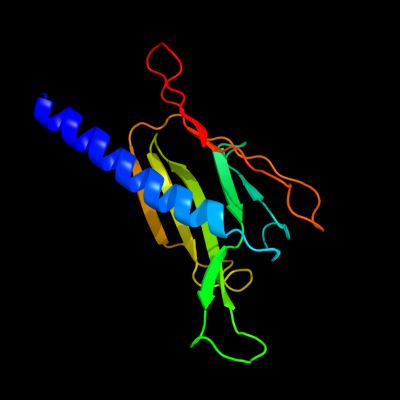
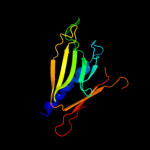
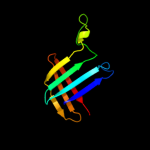
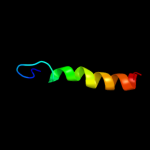
PDB 2ret chain B domain 1
Region: 40 - 176
Aligned: 132
Modelled: 137
Confidence: 98.4%
Identity: 13%
Fold: Pili subunits
Superfamily: Pili subunits
Family: EpsJ-like
Phyre2
| 2 |
|
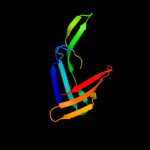
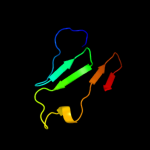
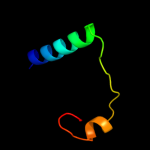
PDB 3ci0 chain J domain 1
Region: 48 - 175
Aligned: 124
Modelled: 126
Confidence: 98.4%
Identity: 11%
Fold: Pili subunits
Superfamily: Pili subunits
Family: EpsJ-like
Phyre2
| 3 |
|
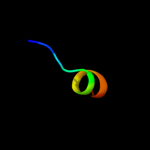
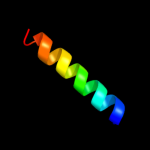
PDB 3sok chain B
Region: 8 - 57
Aligned: 50
Modelled: 50
Confidence: 98.3%
Identity: 18%
PDB header:cell adhesion
Chain: B: PDB Molecule:fimbrial protein;
PDBTitle: dichelobacter nodosus pilin fima
Phyre2
| 4 |
|
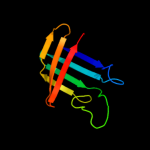
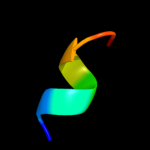
PDB 1oqw chain A
Region: 8 - 60
Aligned: 53
Modelled: 53
Confidence: 98.2%
Identity: 17%
Fold: Pili subunits
Superfamily: Pili subunits
Family: Pilin
Phyre2
| 5 |
|
PDB 2pil chain A
Region: 9 - 61
Aligned: 53
Modelled: 53
Confidence: 98.0%
Identity: 21%
Fold: Pili subunits
Superfamily: Pili subunits
Family: Pilin
Phyre2
| 6 |
|
PDB 3nje chain A
Region: 43 - 175
Aligned: 119
Modelled: 133
Confidence: 97.9%
Identity: 13%
PDB header:protein transport
Chain: A: PDB Molecule:general secretion pathway protein j;
PDBTitle: structure of the minor pseudopilin xcpw from the pseudomonas2 aeruginosa type ii secretion system
Phyre2
| 7 |
|
PDB 2gc9 chain A domain 1
Region: 83 - 164
Aligned: 69
Modelled: 80
Confidence: 38.0%
Identity: 16%
Fold: Lipocalins
Superfamily: Lipocalins
Family: Phenolic acid decarboxylase (PAD)
Phyre2
| 8 |
|
PDB 4a18 chain U
Region: 6 - 16
Aligned: 11
Modelled: 11
Confidence: 29.4%
Identity: 45%
PDB header:ribosome
Chain: U: PDB Molecule:rpl13;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
Phyre2
| 9 |
|
PDB 3u5e chain L
Region: 6 - 16
Aligned: 11
Modelled: 11
Confidence: 27.7%
Identity: 55%
PDB header:ribosome
Chain: L: PDB Molecule:60s ribosomal protein l13-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
Phyre2
| 10 |
|
PDB 3nad chain B
Region: 83 - 164
Aligned: 69
Modelled: 82
Confidence: 23.8%
Identity: 17%
PDB header:lyase
Chain: B: PDB Molecule:ferulate decarboxylase;
PDBTitle: crystal structure of phenolic acid decarboxylase from bacillus pumilus2 ui-670
Phyre2
| 11 |
|
PDB 3nx2 chain B
Region: 83 - 164
Aligned: 69
Modelled: 82
Confidence: 13.8%
Identity: 22%
PDB header:lyase
Chain: B: PDB Molecule:ferulic acid decarboxylase;
PDBTitle: enterobacter sp. px6-4 ferulic acid decarboxylase in complex with2 substrate analogues
Phyre2
| 12 |
|
PDB 3alz chain B
Region: 95 - 146
Aligned: 50
Modelled: 52
Confidence: 12.1%
Identity: 20%
PDB header:viral protein/membrane protein
Chain: B: PDB Molecule:cdw150;
PDBTitle: crystal structure of the measles virus hemagglutinin bound to its2 cellular receptor slam (form i)
Phyre2
| 13 |
|
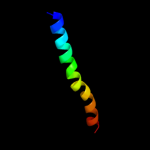
PDB 2kb1 chain A
Region: 8 - 39
Aligned: 32
Modelled: 32
Confidence: 11.5%
Identity: 6%
PDB header:membrane protein
Chain: A: PDB Molecule:wsk3;
PDBTitle: nmr studies of a channel protein without membrane:2 structure and dynamics of water-solubilized kcsa
Phyre2
| 14 |
|
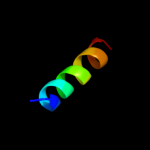
PDB 1r3j chain C
Region: 5 - 40
Aligned: 36
Modelled: 36
Confidence: 10.7%
Identity: 6%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 15 |
|
PDB 3lw5 chain K
Region: 7 - 23
Aligned: 17
Modelled: 17
Confidence: 9.8%
Identity: 24%
PDB header:photosynthesis
Chain: K: PDB Molecule:photosystem i reaction center subunit x psak;
PDBTitle: improved model of plant photosystem i
Phyre2
| 16 |
|
PDB 1jb0 chain K
Region: 4 - 28
Aligned: 25
Modelled: 25
Confidence: 7.9%
Identity: 28%
Fold: Photosystem I reaction center subunit X, PsaK
Superfamily: Photosystem I reaction center subunit X, PsaK
Family: Photosystem I reaction center subunit X, PsaK
Phyre2
| 17 |
|
PDB 1jb0 chain K
Region: 4 - 28
Aligned: 25
Modelled: 25
Confidence: 7.9%
Identity: 28%
PDB header:photosynthesis
Chain: K: PDB Molecule:photosystem 1 reaction centre subunit x;
PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
Phyre2
| 18 |
|
PDB 2hgk chain A domain 1
Region: 48 - 86
Aligned: 39
Modelled: 39
Confidence: 7.6%
Identity: 10%
Fold: Bromodomain-like
Superfamily: YqcC-like
Family: YqcC-like
Phyre2
| 19 |
|
PDB 2axt chain H domain 1
Region: 13 - 33
Aligned: 21
Modelled: 21
Confidence: 6.7%
Identity: 19%
Fold: Single transmembrane helix
Superfamily: Photosystem II 10 kDa phosphoprotein PsbH
Family: PsbH-like
Phyre2
| 20 |
|
PDB 2wsf chain G
Region: 7 - 14
Aligned: 8
Modelled: 8
Confidence: 6.5%
Identity: 50%
PDB header:photosynthesis
Chain: G: PDB Molecule:photosystem i reaction center subunit v,
PDBTitle: improved model of plant photosystem i
Phyre2