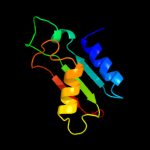
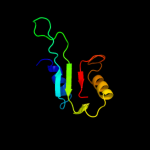
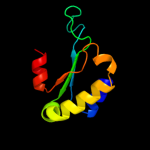
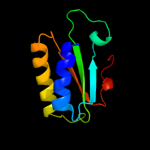
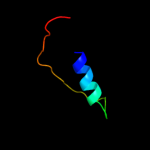
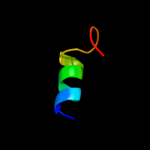
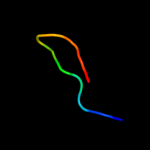
1 c2dhmA_
100.0
100
PDB header: protein bindingChain: A: PDB Molecule: protein bola;PDBTitle: solution structure of the bola protein from escherichia coli
2 c1v60A_
100.0
45
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: riken cdna 1810037g04;PDBTitle: solution structure of bola1 protein from mus musculus
3 c3o2eA_
100.0
35
PDB header: unknown functionChain: A: PDB Molecule: bola-like protein;PDBTitle: crystal structure of a bol-like protein from babesia bovis
4 c2kdnA_
100.0
39
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein pfe0790c;PDBTitle: solution structure of pfe0790c, a putative bola-like2 protein from the protozoan parasite plasmodium falciparum.
5 d1v9ja_
100.0
32
Fold: Alpha-lytic protease prodomain-likeSuperfamily: BolA-likeFamily: BolA-like6 c3tr3A_
100.0
30
PDB header: unknown functionChain: A: PDB Molecule: bola;PDBTitle: structure of a bola protein homologue from coxiella burnetii
7 d1ny8a_
100.0
29
Fold: Alpha-lytic protease prodomain-likeSuperfamily: BolA-likeFamily: BolA-like8 c1xs3A_
100.0
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein xc975;PDBTitle: solution structure analysis of the xc975 protein
9 c2kz0A_
99.9
31
PDB header: transcriptionChain: A: PDB Molecule: bola family protein;PDBTitle: solution structure of a bola protein (ech_0303) from ehrlichia2 chaffeensis. seattle structural genomics center for infectious3 disease target ehcha.10365.a
10 d1ylqa1
96.8
20
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase11 c2rffA_
44.5
15
PDB header: transferaseChain: A: PDB Molecule: putative nucleotidyltransferase;PDBTitle: crystal structure of a putative nucleotidyltransferase2 (np_343093.1) from sulfolobus solfataricus at 1.40 a3 resolution
12 c2hjjA_
18.7
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ykff;PDBTitle: solution nmr structure of protein ykff from escherichia coli.2 northeast structural genomics target er397.
13 d2hjja1
18.7
33
Fold: dsRBD-likeSuperfamily: YcfA/nrd intein domainFamily: YkfF-like14 d1ubdc2
16.3
27
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H215 c2hw4A_
15.8
29
PDB header: structural genomics, hydrolaseChain: A: PDB Molecule: 14 kda phosphohistidine phosphatase;PDBTitle: crystal structure of human phosphohistidine phosphatase
16 d2hw4a1
13.0
29
Fold: PHP14-likeSuperfamily: PHP14-likeFamily: Janus/Ocnus17 c2wu8A_
12.7
15
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: structural studies of phosphoglucose isomerase from2 mycobacterium tuberculosis h37rv
18 d1xhja_
10.3
19
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like19 d1ug8a_
9.7
31
Fold: IF3-likeSuperfamily: R3H domainFamily: R3H domain20 d1x63a1
9.4
36
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain21 d1znfa_
not modelled
9.0
25
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H222 c1znfA_
not modelled
9.0
25
PDB header: zinc finger dna binding domainChain: A: PDB Molecule: 31st zinc finger from xfin;PDBTitle: three-dimensional solution structure of a single zinc2 finger dna-binding domain
23 d1cf7b_
not modelled
8.1
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Cell cycle transcription factor e2f-dp24 d1pvza_
not modelled
8.0
24
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins25 c2yv1A_
not modelled
7.5
9
PDB header: ligaseChain: A: PDB Molecule: succinyl-coa ligase [adp-forming] subunit alpha;PDBTitle: crystal structure of succinyl-coa synthetase alpha chain from2 methanocaldococcus jannaschii dsm 2661
26 c3hjbA_
not modelled
6.3
16
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: 1.5 angstrom crystal structure of glucose-6-phosphate isomerase from2 vibrio cholerae.
27 c3htuE_
not modelled
6.2
25
PDB header: protein transportChain: E: PDB Molecule: vacuolar protein-sorting-associated protein 25;PDBTitle: crystal structure of the human vps25-vps20 subcomplex
28 c3ljkA_
not modelled
6.1
18
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: glucose-6-phosphate isomerase from francisella tularensis.
29 c3ujhB_
not modelled
6.1
20
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of substrate-bound glucose-6-phosphate isomerase2 from toxoplasma gondii
30 d2g7oa1
not modelled
6.0
16
Fold: TraM-likeSuperfamily: TraM-likeFamily: TraM-like31 d1iata_
not modelled
5.9
16
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI32 d1pbya2
not modelled
5.9
33
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 233 c2jsoA_
not modelled
5.6
32
PDB header: signaling proteinChain: A: PDB Molecule: polymyxin resistance protein pmrd;PDBTitle: antimicrobial resistance protein
34 d1u0fa_
not modelled
5.3
16
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI35 d1gzda_
not modelled
5.1
16
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI36 c2btwA_
not modelled
5.1
11
PDB header: transferaseChain: A: PDB Molecule: alr0975 protein;PDBTitle: crystal structure of alr0975