| 1 |
|
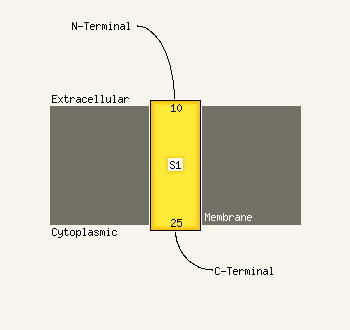
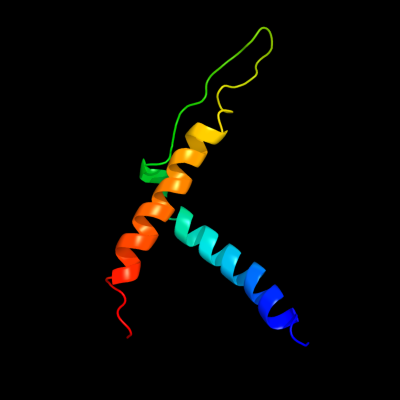
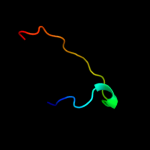
PDB 3hzq chain A
Region: 93 - 164
Aligned: 72
Modelled: 72
Confidence: 26.1%
Identity: 18%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: structure of a tetrameric mscl in an expanded intermediate2 state
Phyre2
| 2 |
|
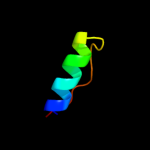
PDB 2vap chain A domain 1
Region: 106 - 129
Aligned: 24
Modelled: 24
Confidence: 15.3%
Identity: 25%
Fold: Tubulin nucleotide-binding domain-like
Superfamily: Tubulin nucleotide-binding domain-like
Family: Tubulin, GTPase domain
Phyre2
| 3 |
|
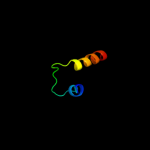
PDB 1ft8 chain E
Region: 27 - 60
Aligned: 34
Modelled: 34
Confidence: 10.7%
Identity: 18%
Fold: Ferredoxin-like
Superfamily: RNA-binding domain, RBD
Family: Non-canonical RBD domain
Phyre2
| 4 |
|
PDB 1ear chain A domain 1
Region: 109 - 131
Aligned: 23
Modelled: 23
Confidence: 9.5%
Identity: 22%
Fold: Urease metallochaperone UreE, N-terminal domain
Superfamily: Urease metallochaperone UreE, N-terminal domain
Family: Urease metallochaperone UreE, N-terminal domain
Phyre2
| 5 |
|
PDB 2d5b chain A domain 1
Region: 105 - 117
Aligned: 13
Modelled: 13
Confidence: 8.5%
Identity: 38%
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
Superfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
Family: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
Phyre2
| 6 |
|
PDB 1w5f chain A domain 1
Region: 106 - 129
Aligned: 24
Modelled: 24
Confidence: 7.6%
Identity: 25%
Fold: Tubulin nucleotide-binding domain-like
Superfamily: Tubulin nucleotide-binding domain-like
Family: Tubulin, GTPase domain
Phyre2
| 7 |
|
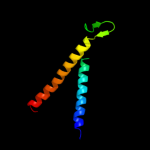
PDB 2oar chain A domain 1
Region: 92 - 164
Aligned: 73
Modelled: 73
Confidence: 7.3%
Identity: 18%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 8 |
|
PDB 1s7b chain A
Region: 78 - 110
Aligned: 33
Modelled: 33
Confidence: 7.0%
Identity: 18%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2