| 1 |
|
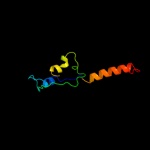
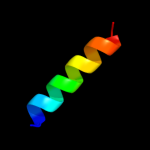
PDB 2lbg chain A
Region: 126 - 137
Aligned: 12
Modelled: 12
Confidence: 16.3%
Identity: 42%
PDB header:membrane protein
Chain: A: PDB Molecule:major prion protein;
PDBTitle: structure of the chr of the prion protein in dpc micelles
Phyre2
| 2 |
|
PDB 3ehw chain A
Region: 194 - 202
Aligned: 9
Modelled: 9
Confidence: 11.6%
Identity: 67%
PDB header:hydrolase
Chain: A: PDB Molecule:dutp pyrophosphatase;
PDBTitle: human dutpase in complex with alpha,beta-imido-dutp and mg2+:2 visualization of the full-length c-termini in all monomers and3 suggestion for an additional metal ion binding site
Phyre2
| 3 |
|
PDB 1gvf chain A
Region: 2 - 84
Aligned: 69
Modelled: 83
Confidence: 11.2%
Identity: 14%
Fold: TIM beta/alpha-barrel
Superfamily: Aldolase
Family: Class II FBP aldolase
Phyre2
| 4 |
|
PDB 1f7r chain A
Region: 194 - 202
Aligned: 9
Modelled: 9
Confidence: 8.8%
Identity: 44%
Fold: beta-clip
Superfamily: dUTPase-like
Family: dUTPase-like
Phyre2
| 5 |
|
PDB 3lyi chain A
Region: 61 - 78
Aligned: 18
Modelled: 18
Confidence: 8.7%
Identity: 17%
PDB header:transcription
Chain: A: PDB Molecule:bromodomain-containing protein 1;
PDBTitle: pwwp domain of human bromodomain-containing protein 1
Phyre2
| 6 |
|
PDB 1dqe chain A
Region: 50 - 72
Aligned: 23
Modelled: 23
Confidence: 6.7%
Identity: 17%
Fold: EF Hand-like
Superfamily: Insect pheromone/odorant-binding proteins
Family: Insect pheromone/odorant-binding proteins
Phyre2
| 7 |
|
PDB 2kxh chain B
Region: 67 - 81
Aligned: 15
Modelled: 15
Confidence: 6.0%
Identity: 40%
PDB header:protein binding
Chain: B: PDB Molecule:peptide of far upstream element-binding protein 1;
PDBTitle: solution structure of the first two rrm domains of fir in the complex2 with fbp nbox peptide
Phyre2
| 8 |
|
PDB 1bik chain A domain 2
Region: 32 - 56
Aligned: 22
Modelled: 25
Confidence: 6.0%
Identity: 18%
Fold: BPTI-like
Superfamily: BPTI-like
Family: Small Kunitz-type inhibitors & BPTI-like toxins
Phyre2
| 9 |
|
PDB 2wcl chain A
Region: 50 - 72
Aligned: 23
Modelled: 23
Confidence: 5.5%
Identity: 17%
PDB header:transport protein
Chain: A: PDB Molecule:general odorant-binding protein 1;
PDBTitle: structure of bmori gobp2 (general odorant binding protein 2)2 with (8e,10z)-hexadecadien-1-ol
Phyre2