| 1 |
|
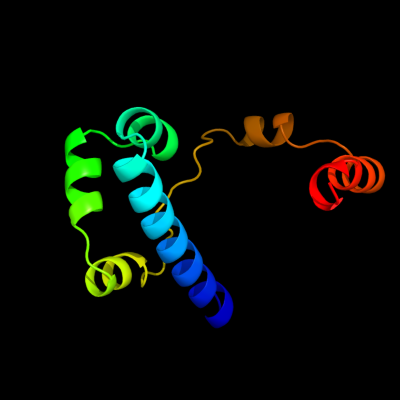
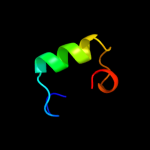
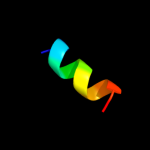
PDB 1g8e chain A
Region: 1 - 98
Aligned: 98
Modelled: 98
Confidence: 100.0%
Identity: 100%
Fold: Flagellar transcriptional activator FlhD
Superfamily: Flagellar transcriptional activator FlhD
Family: Flagellar transcriptional activator FlhD
Phyre2
| 2 |
|
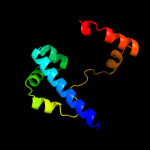
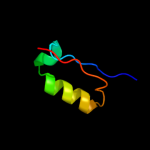
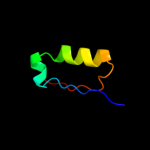
PDB 1g8e chain B
Region: 3 - 82
Aligned: 80
Modelled: 80
Confidence: 100.0%
Identity: 100%
Fold: Flagellar transcriptional activator FlhD
Superfamily: Flagellar transcriptional activator FlhD
Family: Flagellar transcriptional activator FlhD
Phyre2
| 3 |
|
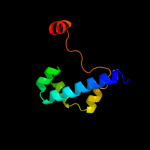
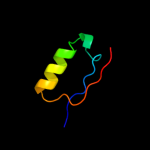
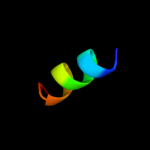
PDB 1xlm chain A
Region: 35 - 71
Aligned: 37
Modelled: 37
Confidence: 18.3%
Identity: 19%
Fold: TIM beta/alpha-barrel
Superfamily: Xylose isomerase-like
Family: Xylose isomerase
Phyre2
| 4 |
|
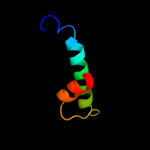
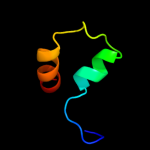
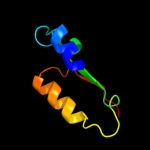
PDB 2kq5 chain A
Region: 48 - 61
Aligned: 14
Modelled: 14
Confidence: 16.9%
Identity: 36%
PDB header:unknown function
Chain: A: PDB Molecule:avirulence protein;
PDBTitle: solution nmr structure of a section of the repeat domain of the type2 iii effector protein ptha
Phyre2
| 5 |
|
PDB 2fx0 chain A domain 2
Region: 7 - 48
Aligned: 42
Modelled: 42
Confidence: 16.0%
Identity: 7%
Fold: Tetracyclin repressor-like, C-terminal domain
Superfamily: Tetracyclin repressor-like, C-terminal domain
Family: Tetracyclin repressor-like, C-terminal domain
Phyre2
| 6 |
|
PDB 1b4u chain A
Region: 21 - 67
Aligned: 47
Modelled: 47
Confidence: 11.7%
Identity: 6%
Fold: LigA subunit of an aromatic-ring-opening dioxygenase LigAB
Superfamily: LigA subunit of an aromatic-ring-opening dioxygenase LigAB
Family: LigA subunit of an aromatic-ring-opening dioxygenase LigAB
Phyre2
| 7 |
|
PDB 3e46 chain A domain 1
Region: 64 - 89
Aligned: 26
Modelled: 26
Confidence: 10.1%
Identity: 23%
Fold: RuvA C-terminal domain-like
Superfamily: UBA-like
Family: UBA domain
Phyre2
| 8 |
|
PDB 1qt1 chain A
Region: 35 - 71
Aligned: 37
Modelled: 37
Confidence: 8.1%
Identity: 22%
Fold: TIM beta/alpha-barrel
Superfamily: Xylose isomerase-like
Family: Xylose isomerase
Phyre2
| 9 |
|
PDB 3m91 chain B
Region: 2 - 13
Aligned: 12
Modelled: 12
Confidence: 8.0%
Identity: 42%
PDB header:hydrolase regulator
Chain: B: PDB Molecule:prokaryotic ubiquitin-like protein pup;
PDBTitle: crystal structure of the prokaryotic ubiquitin-like protein (pup)2 complexed with the amino terminal coiled coil of the mycobacterium3 tuberculosis proteasomal atpase mpa
Phyre2
| 10 |
|
PDB 2glk chain A domain 1
Region: 35 - 71
Aligned: 37
Modelled: 37
Confidence: 7.9%
Identity: 22%
Fold: TIM beta/alpha-barrel
Superfamily: Xylose isomerase-like
Family: Xylose isomerase
Phyre2
| 11 |
|
PDB 1au7 chain A domain 2
Region: 34 - 67
Aligned: 29
Modelled: 34
Confidence: 7.8%
Identity: 24%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: POU-specific domain
Phyre2
| 12 |
|
PDB 3hgf chain A
Region: 7 - 17
Aligned: 11
Modelled: 11
Confidence: 7.6%
Identity: 45%
PDB header:nucleotide binding protein
Chain: A: PDB Molecule:rhoptry protein fragment;
PDBTitle: expression, purification, spectroscopical and2 crystallographical studies of segments of the nucleotide3 binding domain of the reticulocyte binding protein py235 of4 plasmodium yoelii
Phyre2
| 13 |
|
PDB 1dd3 chain A domain 1
Region: 1 - 18
Aligned: 18
Modelled: 18
Confidence: 7.3%
Identity: 28%
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Superfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Family: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Phyre2
| 14 |
|
PDB 1bxc chain A
Region: 35 - 71
Aligned: 37
Modelled: 37
Confidence: 6.7%
Identity: 30%
Fold: TIM beta/alpha-barrel
Superfamily: Xylose isomerase-like
Family: Xylose isomerase
Phyre2
| 15 |
|
PDB 3m9d chain I
Region: 2 - 13
Aligned: 12
Modelled: 12
Confidence: 6.5%
Identity: 42%
PDB header:chaperone
Chain: I: PDB Molecule:prokaryotic ubiquitin-like protein pup;
PDBTitle: crystal structure of the prokaryotic ubiquintin-like protein pup2 complexed with the hexameric proteasomal atpase mpa which includes3 the amino terminal coiled coil domain and the inter domain
Phyre2
| 16 |
|
PDB 2fh0 chain A
Region: 51 - 78
Aligned: 26
Modelled: 28
Confidence: 5.9%
Identity: 23%
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical 16.0 kda protein in abf2-chl12
PDBTitle: nmr ensemble of the yeast saccharomyces cerevisiae protein2 ymr074cp core region
Phyre2
| 17 |
|
PDB 2i56 chain A
Region: 16 - 71
Aligned: 48
Modelled: 56
Confidence: 5.6%
Identity: 15%
PDB header:isomerase, metal-binding protein
Chain: A: PDB Molecule:l-rhamnose isomerase;
PDBTitle: crystal structure of l-rhamnose isomerase from pseudomonas2 stutzeri with l-rhamnose
Phyre2
| 18 |
|
PDB 2e8m chain A
Region: 37 - 57
Aligned: 21
Modelled: 21
Confidence: 5.6%
Identity: 29%
PDB header:signaling protein
Chain: A: PDB Molecule:epidermal growth factor receptor kinase
PDBTitle: solution structure of the c-terminal sam-domain of2 epidermal growth receptor pathway substrate 8
Phyre2
| 19 |
|
PDB 3eq1 chain A
Region: 30 - 92
Aligned: 60
Modelled: 63
Confidence: 5.5%
Identity: 10%
PDB header:transferase
Chain: A: PDB Molecule:porphobilinogen deaminase;
PDBTitle: the crystal structure of human porphobilinogen deaminase at2 2.8a resolution
Phyre2
| 20 |
|
PDB 2avu chain F
Region: 6 - 97
Aligned: 61
Modelled: 61
Confidence: 5.4%
Identity: 31%
PDB header:transcription activator
Chain: F: PDB Molecule:flagellar transcriptional activator flhc;
PDBTitle: structure of the escherichia coli flhdc complex, a2 prokaryotic heteromeric regulator of transcription
Phyre2