| 1 |
|
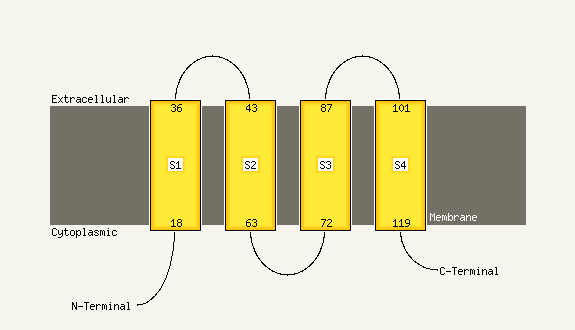
PDB 2axt chain I domain 1
Region: 102 - 127
Aligned: 26
Modelled: 26
Confidence: 16.1%
Identity: 15%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein I, PsbI
Family: PsbI-like
Phyre2
| 2 |
|
PDB 3gia chain A
Region: 9 - 135
Aligned: 126
Modelled: 127
Confidence: 15.5%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 3 |
|
PDB 2dyo chain B
Region: 61 - 70
Aligned: 10
Modelled: 10
Confidence: 12.8%
Identity: 40%
PDB header:protein turnover/protein turnover
Chain: B: PDB Molecule:autophagy protein 16;
PDBTitle: the crystal structure of saccharomyces cerevisiae atg5-2 atg16(1-57) complex
Phyre2
| 4 |
|
PDB 1bzg chain A
Region: 66 - 72
Aligned: 7
Modelled: 7
Confidence: 11.5%
Identity: 71%
PDB header:hormone
Chain: A: PDB Molecule:parathyroid hormone-related protein;
PDBTitle: the solution structure of human parathyroid hormone-related2 protein (1-34) in near-physiological solution, nmr, 303 structures
Phyre2
| 5 |
|
PDB 3zrk chain Y
Region: 62 - 67
Aligned: 6
Modelled: 6
Confidence: 11.2%
Identity: 67%
PDB header:transferase/peptide
Chain: Y: PDB Molecule:proto-oncogene frat1;
PDBTitle: identification of 2-(4-pyridyl)thienopyridinones as gsk-3beta2 inhibitors
Phyre2
| 6 |
|
PDB 1gng chain X
Region: 62 - 67
Aligned: 6
Modelled: 6
Confidence: 10.7%
Identity: 67%
PDB header:transferase
Chain: X: PDB Molecule:frattide;
PDBTitle: glycogen synthase kinase-3 beta (gsk3) complex with frattide2 peptide
Phyre2
| 7 |
|
PDB 1lir chain A
Region: 64 - 71
Aligned: 8
Modelled: 8
Confidence: 10.6%
Identity: 63%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Scorpion toxin-like
Family: Short-chain scorpion toxins
Phyre2
| 8 |
|
PDB 1tqe chain Y
Region: 132 - 145
Aligned: 14
Modelled: 14
Confidence: 8.7%
Identity: 7%
PDB header:transcription/protein binding/dna
Chain: Y: PDB Molecule:histone deacetylase 9;
PDBTitle: mechanism of recruitment of class ii histone deacetylases2 by myocyte enhancer factor-2
Phyre2
| 9 |
|
PDB 1tqe chain X
Region: 132 - 145
Aligned: 14
Modelled: 14
Confidence: 8.7%
Identity: 7%
PDB header:transcription/protein binding/dna
Chain: X: PDB Molecule:histone deacetylase 9;
PDBTitle: mechanism of recruitment of class ii histone deacetylases2 by myocyte enhancer factor-2
Phyre2
| 10 |
|
PDB 1zcz chain A
Region: 128 - 135
Aligned: 8
Modelled: 8
Confidence: 8.5%
Identity: 25%
PDB header:transferase/hydrolase
Chain: A: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of phosphoribosylaminoimidazolecarboxamide2 formyltransferase / imp cyclohydrolase (tm1249) from thermotoga3 maritima at 1.88 a resolution
Phyre2
| 11 |
|
PDB 1jwy chain A domain 1
Region: 129 - 140
Aligned: 12
Modelled: 12
Confidence: 8.4%
Identity: 17%
Fold: SH3-like barrel
Superfamily: Myosin S1 fragment, N-terminal domain
Family: Myosin S1 fragment, N-terminal domain
Phyre2
| 12 |
|
PDB 3skd chain A
Region: 66 - 71
Aligned: 6
Modelled: 6
Confidence: 7.5%
Identity: 67%
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein tthb187;
PDBTitle: crystal structure of the thermus thermophilus cas3 hd domain in the2 presence of ni2+
Phyre2
| 13 |
|
PDB 3bz7 chain A domain 1
Region: 129 - 140
Aligned: 12
Modelled: 12
Confidence: 7.5%
Identity: 17%
Fold: SH3-like barrel
Superfamily: Myosin S1 fragment, N-terminal domain
Family: Myosin S1 fragment, N-terminal domain
Phyre2
| 14 |
|
PDB 2eyq chain A domain 6
Region: 128 - 138
Aligned: 11
Modelled: 11
Confidence: 7.2%
Identity: 9%
Fold: TRCF domain-like
Superfamily: TRCF domain-like
Family: TRCF domain
Phyre2
| 15 |
|
PDB 2ogi chain A
Region: 66 - 71
Aligned: 6
Modelled: 6
Confidence: 6.7%
Identity: 50%
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein sag1661;
PDBTitle: crystal structure of a putative metal dependent phosphohydrolase2 (sag1661) from streptococcus agalactiae serogroup v at 1.85 a3 resolution
Phyre2
| 16 |
|
PDB 2o08 chain B
Region: 66 - 71
Aligned: 6
Modelled: 6
Confidence: 6.3%
Identity: 33%
PDB header:hydrolase
Chain: B: PDB Molecule:bh1327 protein;
PDBTitle: crystal structure of a putative hd superfamily hydrolase (bh1327) from2 bacillus halodurans at 1.90 a resolution
Phyre2
| 17 |
|
PDB 3ccg chain A
Region: 66 - 71
Aligned: 6
Modelled: 6
Confidence: 5.9%
Identity: 33%
PDB header:hydrolase
Chain: A: PDB Molecule:hd superfamily hydrolase;
PDBTitle: crystal structure of predicted hd superfamily hydrolase involved in2 nad metabolism (np_347894.1) from clostridium acetobutylicum at 1.503 a resolution
Phyre2
| 18 |
|
PDB 2crd chain A
Region: 64 - 71
Aligned: 8
Modelled: 8
Confidence: 5.8%
Identity: 50%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Scorpion toxin-like
Family: Short-chain scorpion toxins
Phyre2
| 19 |
|
PDB 2qsr chain A
Region: 128 - 138
Aligned: 11
Modelled: 11
Confidence: 5.8%
Identity: 27%
PDB header:transcription
Chain: A: PDB Molecule:transcription-repair coupling factor;
PDBTitle: crystal structure of c-terminal domain of transcription-repair2 coupling factor
Phyre2
| 20 |
|
PDB 4a1o chain B
Region: 128 - 146
Aligned: 19
Modelled: 19
Confidence: 5.7%
Identity: 16%
PDB header:transferase-hydrolase
Chain: B: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of mycobacterium tuberculosis purh complexed with2 aicar and a novel nucleotide cfair, at 2.48 a resolution.
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|