| 1 |
|
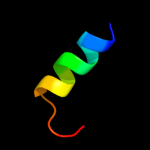
PDB 2haf chain A domain 1
Region: 130 - 139
Aligned: 10
Modelled: 10
Confidence: 17.7%
Identity: 10%
Fold: VC0467-like
Superfamily: VC0467-like
Family: VC0467-like
Phyre2
| 2 |
|
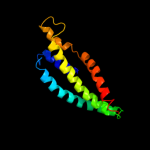
PDB 1abz chain A
Region: 124 - 139
Aligned: 16
Modelled: 16
Confidence: 13.7%
Identity: 25%
PDB header:de novo design
Chain: A: PDB Molecule:alpha-t-alpha;
PDBTitle: alpha-t-alpha, a de novo designed peptide, nmr, 232 structures
Phyre2
| 3 |
|
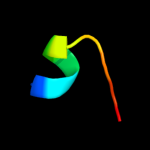
PDB 2yvx chain D
Region: 23 - 179
Aligned: 118
Modelled: 129
Confidence: 13.6%
Identity: 17%
PDB header:transport protein
Chain: D: PDB Molecule:mg2+ transporter mgte;
PDBTitle: crystal structure of magnesium transporter mgte
Phyre2
| 4 |
|
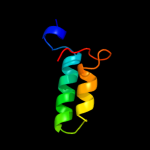
PDB 2aj2 chain A
Region: 130 - 139
Aligned: 10
Modelled: 10
Confidence: 13.1%
Identity: 10%
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical upf0301 protein vc0467;
PDBTitle: x-ray crystal structure of protein vc0467 from vibrio2 cholerae. northeast structural genomics consortium target3 vcr8.
Phyre2
| 5 |
|
PDB 2hv8 chain D
Region: 115 - 133
Aligned: 19
Modelled: 19
Confidence: 13.0%
Identity: 26%
PDB header:protein transport
Chain: D: PDB Molecule:rab11 family-interacting protein 3;
PDBTitle: crystal structure of gtp-bound rab11 in complex with fip3
Phyre2
| 6 |
|
PDB 2iea chain A domain 3
Region: 95 - 139
Aligned: 42
Modelled: 45
Confidence: 11.5%
Identity: 14%
Fold: TK C-terminal domain-like
Superfamily: TK C-terminal domain-like
Family: Transketolase C-terminal domain-like
Phyre2
| 7 |
|
PDB 1z96 chain A domain 1
Region: 125 - 141
Aligned: 17
Modelled: 17
Confidence: 10.1%
Identity: 24%
Fold: RuvA C-terminal domain-like
Superfamily: UBA-like
Family: UBA domain
Phyre2
| 8 |
|
PDB 2hg5 chain D
Region: 147 - 176
Aligned: 30
Modelled: 30
Confidence: 9.9%
Identity: 20%
PDB header:membrane protein
Chain: D: PDB Molecule:kcsa channel;
PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
Phyre2
| 9 |
|
PDB 2la2 chain A
Region: 2 - 30
Aligned: 29
Modelled: 29
Confidence: 8.4%
Identity: 14%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:cecropin;
PDBTitle: solution structure of papiliocin isolated from the swallowtail2 butterfly, papilio xuthus
Phyre2
| 10 |
|
PDB 2kn8 chain A
Region: 121 - 144
Aligned: 24
Modelled: 24
Confidence: 8.4%
Identity: 25%
PDB header:protein binding, dna binding protein
Chain: A: PDB Molecule:dna cleavage and packaging protein large subunit, ul89;
PDBTitle: nmr structure of the c-terminal domain of pul89
Phyre2
| 11 |
|
PDB 1q7t chain A
Region: 119 - 143
Aligned: 25
Modelled: 25
Confidence: 8.3%
Identity: 16%
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein rv1170;
PDBTitle: rv1170 (mshb) from mycobacterium tuberculosis
Phyre2
| 12 |
|
PDB 1g7o chain A domain 1
Region: 124 - 149
Aligned: 26
Modelled: 26
Confidence: 8.3%
Identity: 12%
Fold: GST C-terminal domain-like
Superfamily: GST C-terminal domain-like
Family: Glutathione S-transferase (GST), C-terminal domain
Phyre2
| 13 |
|
PDB 1cf3 chain A
Region: 101 - 124
Aligned: 24
Modelled: 24
Confidence: 8.1%
Identity: 13%
PDB header:oxidoreductase(flavoprotein)
Chain: A: PDB Molecule:protein (glucose oxidase);
PDBTitle: glucose oxidase from apergillus niger
Phyre2
| 14 |
|
PDB 1vma chain A
Region: 227 - 238
Aligned: 12
Modelled: 12
Confidence: 6.9%
Identity: 17%
PDB header:protein transport
Chain: A: PDB Molecule:cell division protein ftsy;
PDBTitle: crystal structure of cell division protein ftsy (tm0570) from2 thermotoga maritima at 1.60 a resolution
Phyre2
| 15 |
|
PDB 2k0b chain X domain 1
Region: 124 - 138
Aligned: 15
Modelled: 15
Confidence: 6.8%
Identity: 40%
Fold: RuvA C-terminal domain-like
Superfamily: UBA-like
Family: UBA domain
Phyre2
| 16 |
|
PDB 1q74 chain A
Region: 119 - 143
Aligned: 25
Modelled: 25
Confidence: 6.6%
Identity: 16%
Fold: LmbE-like
Superfamily: LmbE-like
Family: LmbE-like
Phyre2
| 17 |
|
PDB 2do8 chain A domain 1
Region: 130 - 139
Aligned: 10
Modelled: 10
Confidence: 6.0%
Identity: 20%
Fold: VC0467-like
Superfamily: VC0467-like
Family: VC0467-like
Phyre2
| 18 |
|
PDB 2hjq chain A domain 1
Region: 118 - 142
Aligned: 25
Modelled: 25
Confidence: 5.9%
Identity: 20%
Fold: LEM/SAP HeH motif
Superfamily: Rho N-terminal domain-like
Family: YqbF C-terminal domain-like
Phyre2
| 19 |
|
PDB 2j7p chain A
Region: 227 - 238
Aligned: 12
Modelled: 12
Confidence: 5.7%
Identity: 25%
PDB header:signal recognition
Chain: A: PDB Molecule:signal recognition particle protein;
PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
Phyre2
| 20 |
|
PDB 3dfm chain A
Region: 119 - 139
Aligned: 21
Modelled: 21
Confidence: 5.5%
Identity: 19%
PDB header:hydrolase
Chain: A: PDB Molecule:teicoplanin pseudoaglycone deacetylase orf2;
PDBTitle: the crystal structure of the zinc inhibited form of2 teicoplanin deacetylase orf2
Phyre2
| 21 |
|