| 1 |
|
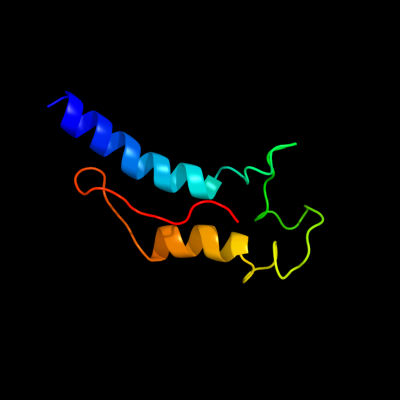
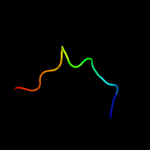
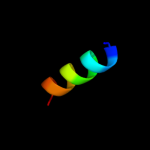
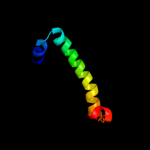
PDB 2kz6 chain A
Region: 39 - 112
Aligned: 64
Modelled: 74
Confidence: 97.6%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of protein cv0426 from chromobacterium violaceum,2 northeast structural genomics consortium (nesg) target cvt2
Phyre2
| 2 |
|
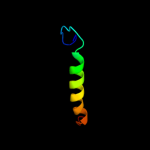
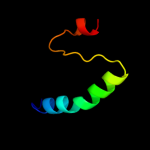
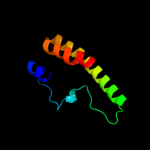
PDB 2kt9 chain A
Region: 21 - 69
Aligned: 49
Modelled: 49
Confidence: 53.5%
Identity: 24%
PDB header:ribosomal protein
Chain: A: PDB Molecule:probable 30s ribosomal protein psrp-3;
PDBTitle: solution nmr structure of probable 30s ribosomal protein2 psrp-3 (ycf65-like protein) from synechocystis sp. (strain3 pcc 6803), northeast structural genomics consortium target4 target sgr46
Phyre2
| 3 |
|
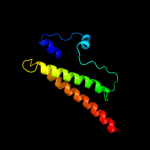
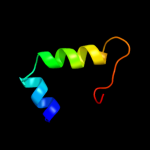
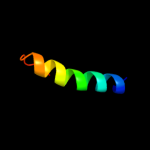
PDB 3o0r chain C
Region: 77 - 105
Aligned: 26
Modelled: 29
Confidence: 37.3%
Identity: 23%
PDB header:immune system/oxidoreductase
Chain: C: PDB Molecule:nitric oxide reductase subunit c;
PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
Phyre2
| 4 |
|
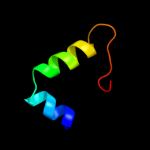
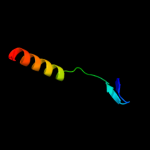
PDB 2oa5 chain A domain 1
Region: 38 - 95
Aligned: 54
Modelled: 58
Confidence: 13.0%
Identity: 19%
Fold: BLRF2-like
Superfamily: BLRF2-like
Family: BLRF2-like
Phyre2
| 5 |
|
PDB 1ew4 chain A
Region: 21 - 35
Aligned: 15
Modelled: 15
Confidence: 11.9%
Identity: 20%
Fold: N domain of copper amine oxidase-like
Superfamily: Frataxin/Nqo15-like
Family: Frataxin-like
Phyre2
| 6 |
|
PDB 2z8n chain B
Region: 19 - 29
Aligned: 11
Modelled: 11
Confidence: 10.9%
Identity: 36%
PDB header:lyase
Chain: B: PDB Molecule:27.5 kda virulence protein;
PDBTitle: structural basis for the catalytic mechanism of phosphothreonine lyase
Phyre2
| 7 |
|
PDB 3ok8 chain A
Region: 29 - 97
Aligned: 69
Modelled: 69
Confidence: 9.0%
Identity: 4%
PDB header:protein binding
Chain: A: PDB Molecule:brain-specific angiogenesis inhibitor 1-associated protein
PDBTitle: i-bar of pinkbar
Phyre2
| 8 |
|
PDB 1b35 chain D
Region: 8 - 21
Aligned: 14
Modelled: 14
Confidence: 7.9%
Identity: 21%
PDB header:virus
Chain: D: PDB Molecule:protein (cricket paralysis virus, vp4);
PDBTitle: cricket paralysis virus (crpv)
Phyre2
| 9 |
|
PDB 2f2a chain C domain 1
Region: 70 - 102
Aligned: 33
Modelled: 33
Confidence: 7.6%
Identity: 24%
Fold: Non-globular all-alpha subunits of globular proteins
Superfamily: Glu-tRNAGln amidotransferase C subunit
Family: Glu-tRNAGln amidotransferase C subunit
Phyre2
| 10 |
|
PDB 2vzb chain A
Region: 2 - 103
Aligned: 99
Modelled: 102
Confidence: 7.6%
Identity: 10%
PDB header:metal transport
Chain: A: PDB Molecule:putative bacterioferritin-related protein;
PDBTitle: a dodecameric thioferritin in the bacterial domain, characterization2 of the bacterioferritin-related protein from bacteroides fragilis
Phyre2
| 11 |
|
PDB 2dii chain A domain 1
Region: 51 - 81
Aligned: 30
Modelled: 31
Confidence: 7.2%
Identity: 7%
Fold: BSD domain-like
Superfamily: BSD domain-like
Family: BSD domain
Phyre2
| 12 |
|
PDB 1nwd chain C
Region: 78 - 91
Aligned: 14
Modelled: 14
Confidence: 7.2%
Identity: 0%
PDB header:binding protein/hydrolase
Chain: C: PDB Molecule:glutamate decarboxylase;
PDBTitle: solution structure of ca2+/calmodulin bound to the c-2 terminal domain of petunia glutamate decarboxylase
Phyre2
| 13 |
|
PDB 1nwd chain B
Region: 78 - 91
Aligned: 14
Modelled: 14
Confidence: 7.2%
Identity: 0%
PDB header:binding protein/hydrolase
Chain: B: PDB Molecule:glutamate decarboxylase;
PDBTitle: solution structure of ca2+/calmodulin bound to the c-2 terminal domain of petunia glutamate decarboxylase
Phyre2
| 14 |
|
PDB 2axp chain A domain 1
Region: 2 - 41
Aligned: 39
Modelled: 40
Confidence: 7.1%
Identity: 28%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: Nucleotide and nucleoside kinases
Phyre2
| 15 |
|
PDB 2dii chain A
Region: 51 - 81
Aligned: 30
Modelled: 31
Confidence: 6.8%
Identity: 7%
PDB header:transcription
Chain: A: PDB Molecule:tfiih basal transcription factor complex p62
PDBTitle: solution structure of the bsd domain of human tfiih basal2 transcription factor complex p62 subunit
Phyre2
| 16 |
|
PDB 2paj chain A domain 1
Region: 27 - 60
Aligned: 34
Modelled: 34
Confidence: 6.8%
Identity: 12%
Fold: Composite domain of metallo-dependent hydrolases
Superfamily: Composite domain of metallo-dependent hydrolases
Family: SAH/MTA deaminase-like
Phyre2
| 17 |
|
PDB 3lvy chain B
Region: 65 - 112
Aligned: 46
Modelled: 48
Confidence: 5.9%
Identity: 4%
PDB header:lyase
Chain: B: PDB Molecule:carboxymuconolactone decarboxylase family;
PDBTitle: crystal structure of carboxymuconolactone decarboxylase2 family protein smu.961 from streptococcus mutans
Phyre2
| 18 |
|
PDB 2d5k chain C
Region: 3 - 82
Aligned: 79
Modelled: 80
Confidence: 5.5%
Identity: 10%
PDB header:metal binding protein
Chain: C: PDB Molecule:dps family protein;
PDBTitle: crystal structure of dps from staphylococcus aureus
Phyre2
| 19 |
|
PDB 1bgv chain A domain 2
Region: 78 - 101
Aligned: 24
Modelled: 24
Confidence: 5.3%
Identity: 13%
Fold: Aminoacid dehydrogenase-like, N-terminal domain
Superfamily: Aminoacid dehydrogenase-like, N-terminal domain
Family: Aminoacid dehydrogenases
Phyre2