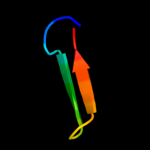
1 d1tiha_
81.2
35
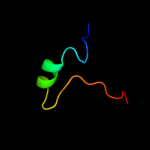
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors2 d1pjua1
80.7
39
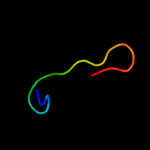
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors3 d1fyba1
80.1
39
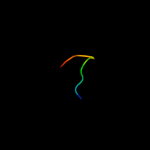
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors4 d4sgbi_
77.6
35
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors5 d1oyvi_
71.6
39
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors6 c1fybA_
69.3
39
PDB header: hydrolase inhibitorChain: A: PDB Molecule: proteinase inhibitor;PDBTitle: solution structure of c1-t1, a two-domain proteinase2 inhibitor derived from the circular precursor protein na-3 propi from nicotiana alata
7 d1ce3a_
50.6
41
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors8 d1pjua2
35.9
45
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors9 c2d3jA_
13.3
40
PDB header: signaling protein inhibitorChain: A: PDB Molecule: wnt inhibitory factor-1;PDBTitle: nmr structure of the wif domain from human wif-1
10 c2e3eA_
12.5
38
PDB header: antimicrobial proteinChain: A: PDB Molecule: defensin, mutant def-bbb;PDBTitle: nmr structure of def-bbb, a mutant of anopheles defensin2 def-aaa
11 d2jnaa1
11.9
30
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like12 c2e2fA_
10.2
40
PDB header: antifungal proteinChain: A: PDB Molecule: diapausin;PDBTitle: solution structure of dsp
13 d1zuea1
9.6
56
Fold: Defensin-likeSuperfamily: Defensin-likeFamily: Defensin14 c1xfeA_
9.5
35
PDB header: lipid transport, endocytosis/exocytosisChain: A: PDB Molecule: low-density lipoprotein receptor;PDBTitle: solution structure of the la7-egfa pair from the ldl2 receptor
15 d2cupa2
9.5
40
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain16 d1e43a1
8.3
55
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain17 d1bgva2
8.2
31
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases18 d1ud2a1
8.2
55
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain19 d2d3na1
8.0
55
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain20 d2gjpa1
7.8
45
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain21 d1eoka_
not modelled
7.3
33
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase22 c1ozbI_
not modelled
6.1
67
PDB header: protein transportChain: I: PDB Molecule: preprotein translocase seca subunit;PDBTitle: crystal structure of secb complexed with seca c-terminus
23 d1ozbi_
not modelled
6.1
67
Fold: Sec-C motifSuperfamily: Sec-C motifFamily: Sec-C motif24 c1ozbJ_
not modelled
6.1
67
PDB header: protein transportChain: J: PDB Molecule: preprotein translocase seca subunit;PDBTitle: crystal structure of secb complexed with seca c-terminus
25 c1d2jA_
not modelled
6.0
26
PDB header: signaling proteinChain: A: PDB Molecule: low-density lipoprotein receptor;PDBTitle: ldl receptor ligand-binding module 6
26 d1hvxa1
not modelled
5.9
55
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain27 c4a1eT_
not modelled
5.9
36
PDB header: ribosomeChain: T: PDB Molecule: rpl24;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
28 d1v9la2
not modelled
5.8
19
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases29 d1g26a_
not modelled
5.7
38
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Granulin repeatFamily: Granulin repeat30 c2i1pA_
not modelled
5.7
43
PDB header: ligand binding proteinChain: A: PDB Molecule: low-density lipoprotein receptor-related proteinPDBTitle: solution structure of the twelfth cysteine-rich ligand-2 binding repeat in rat megalin
31 d1euza2
not modelled
5.6
27
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases32 d1hwxa2
not modelled
5.6
19
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases33 d1tm6a_
not modelled
5.5
67
Fold: Sec-C motifSuperfamily: Sec-C motifFamily: Sec-C motif34 c1tm6A_
not modelled
5.5
67
PDB header: protein transportChain: A: PDB Molecule: preprotein translocase seca subunit;PDBTitle: nmr structure of the free zinc binding c-terminal domain of2 seca