| 1 |
|
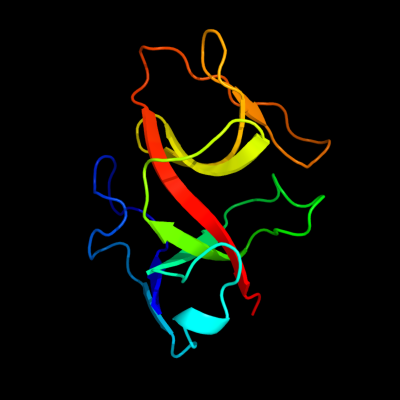
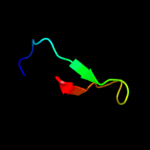
PDB 1jhf chain A domain 2
Region: 15 - 137
Aligned: 121
Modelled: 123
Confidence: 100.0%
Identity: 33%
Fold: LexA/Signal peptidase
Superfamily: LexA/Signal peptidase
Family: LexA-related
Phyre2
| 2 |
|
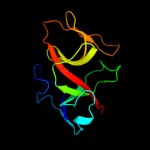
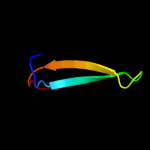
PDB 1umu chain A
Region: 32 - 136
Aligned: 105
Modelled: 105
Confidence: 100.0%
Identity: 98%
Fold: LexA/Signal peptidase
Superfamily: LexA/Signal peptidase
Family: LexA-related
Phyre2
| 3 |
|
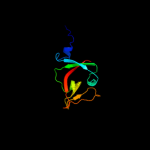
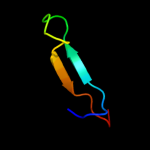
PDB 3k2z chain A
Region: 14 - 138
Aligned: 123
Modelled: 125
Confidence: 99.9%
Identity: 33%
PDB header:hydrolase
Chain: A: PDB Molecule:lexa repressor;
PDBTitle: crystal structure of a lexa protein from thermotoga maritima
Phyre2
| 4 |
|
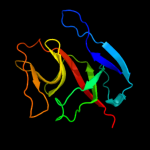
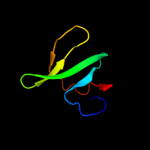
PDB 1jhh chain B
Region: 15 - 137
Aligned: 114
Modelled: 123
Confidence: 99.9%
Identity: 32%
PDB header:hydrolase
Chain: B: PDB Molecule:lexa repressor;
PDBTitle: lexa s119a mutant
Phyre2
| 5 |
|
PDB 2hnf chain A
Region: 10 - 134
Aligned: 122
Modelled: 125
Confidence: 99.9%
Identity: 29%
PDB header:viral protein
Chain: A: PDB Molecule:repressor protein ci101-229dm-k192a;
PDBTitle: structure of a hyper-cleavable monomeric fragment of phage2 lambda repressor containing the cleavage site region
Phyre2
| 6 |
|
PDB 1f39 chain A
Region: 49 - 136
Aligned: 86
Modelled: 88
Confidence: 99.8%
Identity: 34%
Fold: LexA/Signal peptidase
Superfamily: LexA/Signal peptidase
Family: LexA-related
Phyre2
| 7 |
|
PDB 3bdn chain B
Region: 6 - 135
Aligned: 125
Modelled: 130
Confidence: 99.7%
Identity: 32%
PDB header:transcription/dna
Chain: B: PDB Molecule:lambda repressor;
PDBTitle: crystal structure of the lambda repressor
Phyre2
| 8 |
|
PDB 2fjr chain B
Region: 3 - 138
Aligned: 114
Modelled: 125
Confidence: 99.6%
Identity: 10%
PDB header:transcription regulator
Chain: B: PDB Molecule:repressor protein ci;
PDBTitle: crystal structure of bacteriophage 186
Phyre2
| 9 |
|
PDB 2fkd chain K
Region: 68 - 138
Aligned: 66
Modelled: 71
Confidence: 98.7%
Identity: 18%
PDB header:transcription regulator
Chain: K: PDB Molecule:repressor protein ci;
PDBTitle: crystal structure of the c-terminal domain of bacteriophage2 186 repressor
Phyre2
| 10 |
|
PDB 1b12 chain A
Region: 52 - 113
Aligned: 62
Modelled: 62
Confidence: 97.5%
Identity: 21%
Fold: LexA/Signal peptidase
Superfamily: LexA/Signal peptidase
Family: Type 1 signal peptidase
Phyre2
| 11 |
|
PDB 1nnx chain A
Region: 86 - 131
Aligned: 45
Modelled: 46
Confidence: 15.0%
Identity: 18%
Fold: OB-fold
Superfamily: Hypothetical protein YgiW
Family: Hypothetical protein YgiW
Phyre2
| 12 |
|
PDB 1hr0 chain W
Region: 66 - 87
Aligned: 22
Modelled: 22
Confidence: 14.9%
Identity: 32%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 13 |
|
PDB 1jt8 chain A
Region: 66 - 93
Aligned: 28
Modelled: 28
Confidence: 14.6%
Identity: 21%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 14 |
|
PDB 1d7q chain A
Region: 66 - 93
Aligned: 28
Modelled: 28
Confidence: 14.0%
Identity: 18%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 15 |
|
PDB 2uwq chain A
Region: 58 - 66
Aligned: 8
Modelled: 9
Confidence: 10.5%
Identity: 38%
PDB header:apoptosis
Chain: A: PDB Molecule:apoptosis-stimulating of p53 protein 2;
PDBTitle: solution structure of aspp2 n-terminus
Phyre2
| 16 |
|
PDB 2p84 chain A domain 1
Region: 60 - 129
Aligned: 55
Modelled: 55
Confidence: 10.1%
Identity: 22%
Fold: YopX-like
Superfamily: YopX-like
Family: YopX-like
Phyre2
| 17 |
|
PDB 1ah9 chain A
Region: 66 - 87
Aligned: 22
Modelled: 22
Confidence: 9.8%
Identity: 32%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 18 |
|
PDB 2oqk chain A
Region: 66 - 87
Aligned: 22
Modelled: 22
Confidence: 9.1%
Identity: 27%
PDB header:translation
Chain: A: PDB Molecule:putative translation initiation factor eif-1a;
PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
Phyre2
| 19 |
|
PDB 2zpm chain A
Region: 56 - 116
Aligned: 61
Modelled: 61
Confidence: 7.5%
Identity: 11%
PDB header:hydrolase
Chain: A: PDB Molecule:regulator of sigma e protease;
PDBTitle: crystal structure analysis of pdz domain b
Phyre2
| 20 |
|
PDB 1zvp chain A domain 1
Region: 61 - 74
Aligned: 14
Modelled: 14
Confidence: 5.8%
Identity: 14%
Fold: Ferredoxin-like
Superfamily: ACT-like
Family: VC0802-like
Phyre2
| 21 |
|