| 1 |
|
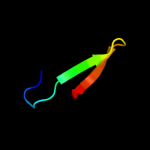
PDB 3m5b chain A
Region: 25 - 42
Aligned: 18
Modelled: 18
Confidence: 18.0%
Identity: 28%
PDB header:transcription
Chain: A: PDB Molecule:zinc finger and btb domain-containing protein 32;
PDBTitle: crystal structure of the btb domain from fazf/zbtb32
Phyre2
| 2 |
|
PDB 2km6 chain A
Region: 15 - 30
Aligned: 16
Modelled: 16
Confidence: 10.2%
Identity: 13%
PDB header:signaling protein, protein binding
Chain: A: PDB Molecule:nacht, lrr and pyd domains-containing protein 7;
PDBTitle: nmr structure of the nlrp7 pyrin domain
Phyre2
| 3 |
|
PDB 1vlf chain N domain 1
Region: 30 - 42
Aligned: 13
Modelled: 13
Confidence: 9.9%
Identity: 38%
Fold: Prealbumin-like
Superfamily: Cna protein B-type domain
Family: Cna protein B-type domain
Phyre2
| 4 |
|
PDB 2qqr chain A domain 1
Region: 23 - 37
Aligned: 15
Modelled: 15
Confidence: 7.9%
Identity: 53%
Fold: SH3-like barrel
Superfamily: Tudor/PWWP/MBT
Family: Tudor domain
Phyre2
| 5 |
|
PDB 2vty chain A
Region: 16 - 25
Aligned: 10
Modelled: 10
Confidence: 6.5%
Identity: 60%
PDB header:apoptosis
Chain: A: PDB Molecule:protein f1;
PDBTitle: vaccinia virus anti-apoptotic f1l is a novel bcl-2-like2 domain swapped dimer
Phyre2
| 6 |
|
PDB 1khi chain A domain 2
Region: 25 - 45
Aligned: 21
Modelled: 21
Confidence: 5.7%
Identity: 29%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 7 |
|
PDB 3qii chain A
Region: 23 - 41
Aligned: 19
Modelled: 19
Confidence: 5.5%
Identity: 21%
PDB header:transcription regulator
Chain: A: PDB Molecule:phd finger protein 20;
PDBTitle: crystal structure of tudor domain 2 of human phd finger protein 20
Phyre2
| 8 |
|
PDB 3arc chain L
Region: 5 - 21
Aligned: 17
Modelled: 17
Confidence: 5.5%
Identity: 35%
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 9 |
|
PDB 1pn5 chain A domain 1
Region: 15 - 30
Aligned: 16
Modelled: 16
Confidence: 5.4%
Identity: 13%
Fold: DEATH domain
Superfamily: DEATH domain
Family: Pyrin domain, PYD
Phyre2
| 10 |
|
PDB 1pn5 chain A
Region: 15 - 30
Aligned: 16
Modelled: 16
Confidence: 5.4%
Identity: 13%
PDB header:apoptosis
Chain: A: PDB Molecule:nacht-, lrr- and pyd-containing protein 2;
PDBTitle: nmr structure of the nalp1 pyrin domain (pyd)
Phyre2
| 11 |
|
PDB 2zxe chain B
Region: 5 - 22
Aligned: 18
Modelled: 18
Confidence: 5.1%
Identity: 28%
PDB header:hydrolase/transport protein
Chain: B: PDB Molecule:na+,k+-atpase beta subunit;
PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
Phyre2