| 1 |
|
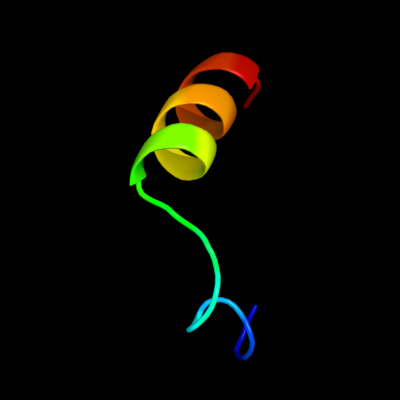
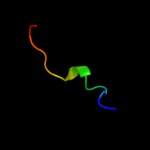
PDB 2isy chain A domain 2
Region: 35 - 53
Aligned: 19
Modelled: 19
Confidence: 10.9%
Identity: 32%
Fold: Iron-dependent repressor protein, dimerization domain
Superfamily: Iron-dependent repressor protein, dimerization domain
Family: Iron-dependent repressor protein, dimerization domain
Phyre2
| 2 |
|
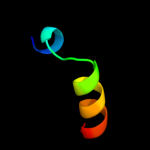
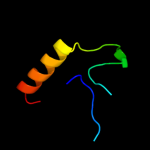
PDB 2ha9 chain A domain 1
Region: 24 - 55
Aligned: 31
Modelled: 32
Confidence: 7.7%
Identity: 23%
Fold: PFL-like glycyl radical enzymes
Superfamily: PFL-like glycyl radical enzymes
Family: SP0239-like
Phyre2
| 3 |
|
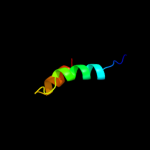
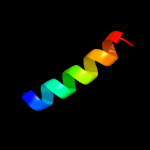
PDB 1h3g chain A domain 1
Region: 14 - 39
Aligned: 26
Modelled: 25
Confidence: 7.3%
Identity: 12%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: E-set domains of sugar-utilizing enzymes
Phyre2
| 4 |
|
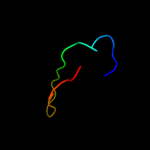
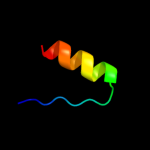
PDB 2p87 chain A
Region: 54 - 68
Aligned: 14
Modelled: 15
Confidence: 7.2%
Identity: 14%
PDB header:splicing
Chain: A: PDB Molecule:pre-mrna-splicing factor prp8;
PDBTitle: crystal structure of the c-terminal domain of c. elegans2 pre-mrna splicing factor prp8
Phyre2
| 5 |
|
PDB 1xb4 chain A domain 1
Region: 21 - 58
Aligned: 38
Modelled: 38
Confidence: 6.5%
Identity: 16%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Vacuolar sorting protein domain
Phyre2
| 6 |
|
PDB 1iow chain A domain 1
Region: 38 - 55
Aligned: 18
Modelled: 18
Confidence: 5.6%
Identity: 39%
Fold: PreATP-grasp domain
Superfamily: PreATP-grasp domain
Family: D-Alanine ligase N-terminal domain
Phyre2
| 7 |
|
PDB 3gpq chain A
Region: 31 - 50
Aligned: 20
Modelled: 20
Confidence: 5.2%
Identity: 35%
PDB header:viral protein/rna
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: crystal structure of macro domain of chikungunya virus in complex with2 rna
Phyre2
| 8 |
|
PDB 3cuq chain D
Region: 37 - 58
Aligned: 22
Modelled: 22
Confidence: 5.2%
Identity: 23%
PDB header:protein transport
Chain: D: PDB Molecule:vacuolar protein-sorting-associated protein 25;
PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
Phyre2