1 c3c1qA_
99.9
26
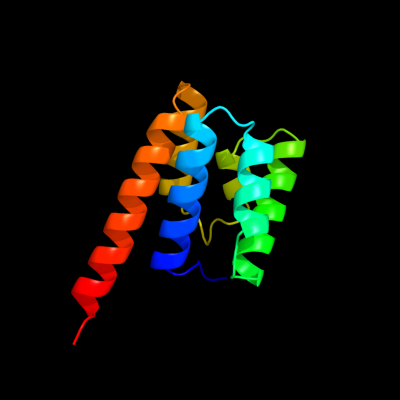
PDB header: transport proteinChain: A: PDB Molecule: general secretion pathway protein f;PDBTitle: the three-dimensional structure of the cytoplasmic domains of epsf2 from the type 2 secretion system of vibrio cholerae
2 c2whnA_
99.8
31
PDB header: protein transportChain: A: PDB Molecule: pilus assembly protein pilc;PDBTitle: n-terminal domain from the pilc type iv pilus biogenesis2 protein
3 c3i3aC_
54.4
13
PDB header: transferaseChain: C: PDB Molecule: acyl-[acyl-carrier-protein]--udp-n-PDBTitle: structural basis for the sugar nucleotide and acyl chain2 selectivity of leptospira interrogans lpxa
4 c2bbjB_
48.5
18
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
5 d3cuma1
47.6
13
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: Hydroxyisobutyrate and 6-phosphogluconate dehydrogenase domain6 c3c8mA_
45.1
14
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: crystal structure of homoserine dehydrogenase from thermoplasma2 volcanium
7 c2ejwB_
41.4
20
PDB header: oxidoreductaseChain: B: PDB Molecule: homoserine dehydrogenase;PDBTitle: homoserine dehydrogenase from thermus thermophilus hb8
8 d2jf2a1
35.6
16
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: UDP N-acetylglucosamine acyltransferase9 c2d1lA_
35.1
11
PDB header: protein bindingChain: A: PDB Molecule: metastasis suppressor protein 1;PDBTitle: structure of f-actin binding domain imd of mim (missing in metastasis)
10 d1y2oa1
31.7
7
Fold: BAR/IMD domain-likeSuperfamily: BAR/IMD domain-likeFamily: IMD domain11 c3ok8A_
29.4
12
PDB header: protein bindingChain: A: PDB Molecule: brain-specific angiogenesis inhibitor 1-associated proteinPDBTitle: i-bar of pinkbar
12 c2c5qE_
28.0
22
PDB header: structural genomics,unknown functionChain: E: PDB Molecule: rraa-like protein yer010c;PDBTitle: crystal structure of yeast yer010cp
13 d2ezla_
23.5
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain14 c3iugA_
19.0
17
PDB header: splicingChain: A: PDB Molecule: rho/cdc42/rac gtpase-activating protein rics;PDBTitle: crystal structure of the rhogap domain of rics
15 c3ipdB_
18.0
12
PDB header: exocytosisChain: B: PDB Molecule: syntaxin-1a;PDBTitle: helical extension of the neuronal snare complex into the2 membrane, spacegroup i 21 21 21
16 d1j2za_
17.7
11
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: UDP N-acetylglucosamine acyltransferase17 c3r0sA_
17.6
18
PDB header: transferaseChain: A: PDB Molecule: acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-PDBTitle: udp-n-acetylglucosamine acyltransferase from campylobacter jejuni
18 c2jmlA_
15.9
19
PDB header: transcriptionChain: A: PDB Molecule: dna binding domain/transcriptional regulator;PDBTitle: solution structure of the n-terminal domain of cara repressor
19 c3ingA_
15.3
12
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: crystal structure of homoserine dehydrogenase (np_394635.1) from2 thermoplasma acidophilum at 1.95 a resolution
20 c3r1fO_
13.5
20
PDB header: transcriptionChain: O: PDB Molecule: esx-1 secretion-associated regulator espr;PDBTitle: crystal structure of a key regulator of virulence in mycobacterium2 tuberculosis
21 c3k4iC_
not modelled
13.1
10
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein pspto_3204 from2 pseudomonas syringae pv. tomato str. dc3000
22 c2pmzN_
not modelled
13.1
10
PDB header: translation, transferaseChain: N: PDB Molecule: dna-directed rna polymerase subunit n;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
23 c3byiA_
not modelled
12.7
13
PDB header: signaling proteinChain: A: PDB Molecule: rho gtpase activating protein 15;PDBTitle: crystal structure of human rho gtpase activating protein 15 (arhgap15)
24 c3do5A_
not modelled
12.3
5
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: crystal structure of putative homoserine dehydrogenase (np_069768.1)2 from archaeoglobus fulgidus at 2.20 a resolution
25 c3kuqA_
not modelled
12.2
11
PDB header: hydrolase activatorChain: A: PDB Molecule: rho gtpase-activating protein 7;PDBTitle: crystal structure of the dlc1 rhogap domain
26 c3neuA_
not modelled
11.2
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin1836 protein;PDBTitle: the crystal structure of a functionally-unknown protein lin1836 from2 listeria innocua clip11262
27 d1xa6a1
not modelled
10.6
11
Fold: GTPase activation domain, GAPSuperfamily: GTPase activation domain, GAPFamily: BCR-homology GTPase activation domain (BH-domain)28 c3gndC_
not modelled
10.1
9
PDB header: lyaseChain: C: PDB Molecule: aldolase lsrf;PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
29 d1i4ja_
not modelled
9.0
15
Fold: Ribosomal protein L22Superfamily: Ribosomal protein L22Family: Ribosomal protein L2230 d1e8ob_
not modelled
8.4
14
Fold: Signal recognition particle alu RNA binding heterodimer, SRP9/14Superfamily: Signal recognition particle alu RNA binding heterodimer, SRP9/14Family: Signal recognition particle alu RNA binding heterodimer, SRP9/1431 d1jnsa_
not modelled
7.7
19
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase32 c1xa6A_
not modelled
7.7
11
PDB header: signaling proteinChain: A: PDB Molecule: beta2-chimaerin;PDBTitle: crystal structure of the human beta2-chimaerin
33 d1wn0a1
not modelled
7.3
19
Fold: Four-helical up-and-down bundleSuperfamily: Histidine-containing phosphotransfer domain, HPT domainFamily: Phosphorelay protein-like34 c3kc2A_
not modelled
7.2
19
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein ykr070w;PDBTitle: crystal structure of mitochondrial had-like phosphatase from2 saccharomyces cerevisiae
35 d2gyqa1
not modelled
7.1
18
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: YciF-like36 c1zk6A_
not modelled
7.1
15
PDB header: isomeraseChain: A: PDB Molecule: foldase protein prsa;PDBTitle: nmr solution structure of b. subtilis prsa ppiase
37 d1vq3a_
not modelled
7.0
13
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase38 c2zhhA_
not modelled
6.9
15
PDB header: transcriptionChain: A: PDB Molecule: redox-sensitive transcriptional activator soxr;PDBTitle: crystal structure of soxr
39 c3gpkA_
not modelled
6.5
28
PDB header: isomeraseChain: A: PDB Molecule: ppic-type peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of ppic-type peptidyl-prolyl cis-trans isomerase2 domain at 1.55a resolution.
40 d2gs4a1
not modelled
6.5
20
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: YciF-like41 c2ftcM_
not modelled
6.5
19
PDB header: ribosomeChain: M: PDB Molecule: mitochondrial ribosomal protein l22 isoform a;PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
42 c3ibqA_
not modelled
6.5
11
PDB header: transferaseChain: A: PDB Molecule: pyridoxal kinase;PDBTitle: crystal structure of pyridoxal kinase from lactobacillus2 plantarum in complex with atp
43 c1sneB_
not modelled
6.5
46
PDB header: de novo proteinChain: B: PDB Molecule: tetrameric beta-beta-alpha mini-protein;PDBTitle: an oligomeric domain-swapped beta-beta-alpha mini-protein
44 c1sneA_
not modelled
6.5
46
PDB header: de novo proteinChain: A: PDB Molecule: tetrameric beta-beta-alpha mini-protein;PDBTitle: an oligomeric domain-swapped beta-beta-alpha mini-protein
45 c2hx1D_
not modelled
6.5
8
PDB header: hydrolaseChain: D: PDB Molecule: predicted sugar phosphatases of the hadPDBTitle: crystal structure of possible sugar phosphatase, had2 superfamily (zp_00311070.1) from cytophaga hutchinsonii3 atcc 33406 at 2.10 a resolution
46 d1914a2
not modelled
6.4
14
Fold: Signal recognition particle alu RNA binding heterodimer, SRP9/14Superfamily: Signal recognition particle alu RNA binding heterodimer, SRP9/14Family: Signal recognition particle alu RNA binding heterodimer, SRP9/1447 c1ebuA_
not modelled
6.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: homoserine dehydrogenase complex with nad analogue and l-2 homoserine
48 d1gtda_
not modelled
6.0
4
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase49 c2eelA_
not modelled
5.9
17
PDB header: apoptosisChain: A: PDB Molecule: cell death activator cide-a;PDBTitle: solution structure of the cide-n domain of human cell death2 activator cide-a
50 c2i7aA_
not modelled
5.9
12
PDB header: hydrolaseChain: A: PDB Molecule: calpain 13;PDBTitle: domain iv of human calpain 13
51 c2qjhH_
not modelled
5.9
10
PDB header: lyaseChain: H: PDB Molecule: putative aldolase mj0400;PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
52 c2zw2B_
not modelled
5.9
8
PDB header: ligaseChain: B: PDB Molecule: putative uncharacterized protein sts178;PDBTitle: crystal structure of formylglycinamide ribonucleotide amidotransferase2 iii from sulfolobus tokodaii (stpurs)
53 c2rqsA_
not modelled
5.8
20
PDB header: isomeraseChain: A: PDB Molecule: parvulin-like peptidyl-prolyl isomerase;PDBTitle: 3d structure of pin from the psychrophilic archeon cenarcheaum2 symbiosum (cspin)
54 d1hara_
not modelled
5.7
13
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: Reverse transcriptase55 d1oqwa_
not modelled
5.5
11
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin56 d1d4ba_
not modelled
5.5
21
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain57 d1eqfa1
not modelled
5.5
6
Fold: Bromodomain-likeSuperfamily: BromodomainFamily: Bromodomain58 c2kncA_
not modelled
5.5
17
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
59 d1c9fa_
not modelled
5.4
17
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain60 c1zzwA_
not modelled
5.4
15
PDB header: hydrolaseChain: A: PDB Molecule: dual specificity protein phosphatase 10;PDBTitle: crystal structure of catalytic domain of human map kinase2 phosphatase 5
61 d2pila_
not modelled
5.4
21
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin62 c3q6aH_
not modelled
5.4
19
PDB header: structure genomics, unknown functionChain: H: PDB Molecule: uncharacterized protein;PDBTitle: x-ray crystal structure of the protein ssp2350 from staphylococcus2 saprophyticus, northeast structural genomics consortium target syr116
63 d1ibxa_
not modelled
5.3
10
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain64 d2pv2a1
not modelled
5.3
10
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase65 c2kgjA_
not modelled
5.2
16
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase d;PDBTitle: solution structure of parvulin domain of ppid from e.coli
66 d2cbia1
not modelled
5.1
16
Fold: Hyaluronidase domain-likeSuperfamily: Hyaluronidase post-catalytic domain-likeFamily: Hyaluronidase post-catalytic domain-like67 c1zrtD_
not modelled
5.1
4
PDB header: oxidoreductase/metal transportChain: D: PDB Molecule: cytochrome c1;PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
68 c3hefB_
not modelled
5.1
12
PDB header: viral proteinChain: B: PDB Molecule: gene 1 protein;PDBTitle: crystal structure of the bacteriophage sf6 terminase small2 subunit
69 c3nmeA_
not modelled
5.0
17
PDB header: hydrolaseChain: A: PDB Molecule: sex4 glucan phosphatase;PDBTitle: structure of a plant phosphatase