| 1 |
|
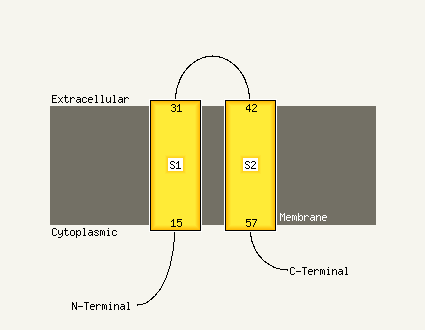
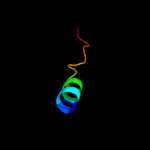
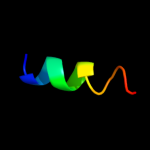
PDB 2knc chain A
Region: 45 - 70
Aligned: 26
Modelled: 26
Confidence: 63.5%
Identity: 27%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 2 |
|
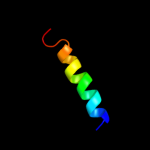
PDB 1dq3 chain A domain 3
Region: 49 - 58
Aligned: 10
Modelled: 10
Confidence: 26.6%
Identity: 50%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Intein endonuclease
Phyre2
| 3 |
|
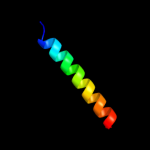
PDB 1m45 chain B
Region: 54 - 62
Aligned: 9
Modelled: 9
Confidence: 22.4%
Identity: 56%
PDB header:cell cycle protein
Chain: B: PDB Molecule:iq2 motif from myo2p, a class v myosin;
PDBTitle: crystal structure of mlc1p bound to iq2 of myo2p, a class v2 myosin
Phyre2
| 4 |
|
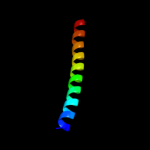
PDB 1jb0 chain I
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 17.1%
Identity: 31%
Fold: Single transmembrane helix
Superfamily: Subunit VIII of photosystem I reaction centre, PsaI
Family: Subunit VIII of photosystem I reaction centre, PsaI
Phyre2
| 5 |
|
PDB 1n2d chain C
Region: 54 - 62
Aligned: 9
Modelled: 9
Confidence: 15.0%
Identity: 56%
PDB header:cell cycle
Chain: C: PDB Molecule:iq2 and iq3 motifs from myo2p, a class v myosin;
PDBTitle: ternary complex of mlc1p bound to iq2 and iq3 of myo2p, a2 class v myosin
Phyre2
| 6 |
|
PDB 2k1a chain A
Region: 45 - 63
Aligned: 19
Modelled: 19
Confidence: 13.3%
Identity: 32%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
Phyre2
| 7 |
|
PDB 2k9y chain B
Region: 42 - 62
Aligned: 21
Modelled: 21
Confidence: 12.7%
Identity: 38%
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 8 |
|
PDB 1vj1 chain A domain 1
Region: 37 - 43
Aligned: 7
Modelled: 7
Confidence: 12.5%
Identity: 57%
Fold: GroES-like
Superfamily: GroES-like
Family: Alcohol dehydrogenase-like, N-terminal domain
Phyre2
| 9 |
|
PDB 2k9y chain A
Region: 42 - 62
Aligned: 21
Modelled: 21
Confidence: 12.4%
Identity: 38%
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 10 |
|
PDB 1bhb chain A
Region: 8 - 38
Aligned: 26
Modelled: 26
Confidence: 9.6%
Identity: 38%
PDB header:photoreceptor
Chain: A: PDB Molecule:bacteriorhodopsin;
PDBTitle: three-dimensional structure of (1-71) bacterioopsin2 solubilized in methanol-chloroform and sds micelles3 determined by 15n-1h heteronuclear nmr spectroscopy
Phyre2
| 11 |
|
PDB 2rcr chain H domain 2
Region: 49 - 64
Aligned: 16
Modelled: 16
Confidence: 8.9%
Identity: 56%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 12 |
|
PDB 2rlw chain A
Region: 42 - 51
Aligned: 10
Modelled: 10
Confidence: 8.8%
Identity: 70%
PDB header:toxin
Chain: A: PDB Molecule:plnf;
PDBTitle: three-dimensional structure of the two peptides that2 constitute the two-peptide bacteriocin plantaracin ef
Phyre2
| 13 |
|
PDB 2bs2 chain C domain 1
Region: 4 - 39
Aligned: 36
Modelled: 36
Confidence: 8.2%
Identity: 19%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Fumarate reductase respiratory complex cytochrome b subunit, FrdC
Phyre2
| 14 |
|
PDB 2voy chain E
Region: 16 - 29
Aligned: 14
Modelled: 14
Confidence: 8.1%
Identity: 57%
PDB header:hydrolase
Chain: E: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 15 |
|
PDB 1mow chain G
Region: 50 - 58
Aligned: 9
Modelled: 9
Confidence: 7.8%
Identity: 67%
PDB header:hydrolase/dna
Chain: G: PDB Molecule:chimera of homing endonuclease i-dmoi and dna endonuclease
PDBTitle: e-drei
Phyre2
| 16 |
|
PDB 2i5n chain H
Region: 49 - 65
Aligned: 17
Modelled: 17
Confidence: 6.7%
Identity: 24%
PDB header:photosynthesis
Chain: H: PDB Molecule:reaction center protein h chain;
PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
Phyre2
| 17 |
|
PDB 2ex5 chain B
Region: 52 - 58
Aligned: 7
Modelled: 7
Confidence: 6.4%
Identity: 43%
PDB header:hydrolase/dna
Chain: B: PDB Molecule:dna endonuclease i-ceui;
PDBTitle: group i intron-encoded homing endonuclease i-ceui complexed2 with dna
Phyre2
| 18 |
|
PDB 3mx7 chain A
Region: 7 - 13
Aligned: 7
Modelled: 7
Confidence: 6.3%
Identity: 43%
PDB header:apoptosis
Chain: A: PDB Molecule:fas apoptotic inhibitory molecule 1;
PDBTitle: crystal structure analysis of human faim-ntd
Phyre2
| 19 |
|
PDB 1af5 chain A
Region: 53 - 58
Aligned: 6
Modelled: 6
Confidence: 5.8%
Identity: 50%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Group I mobile intron endonuclease
Phyre2
| 20 |
|
PDB 1m5x chain A
Region: 53 - 58
Aligned: 6
Modelled: 6
Confidence: 5.7%
Identity: 33%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Group I mobile intron endonuclease
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|