| 1 |
|
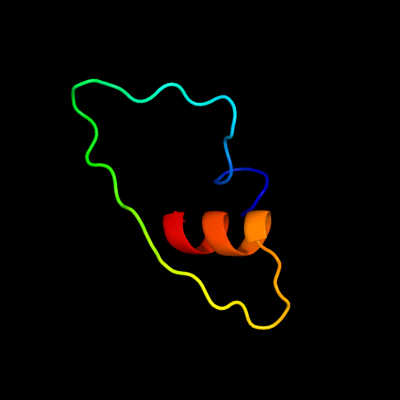
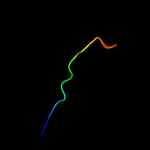
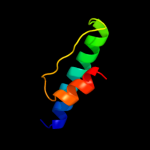
PDB 1ej7 chain L domain 2
Region: 18 - 56
Aligned: 39
Modelled: 39
Confidence: 19.6%
Identity: 15%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 2 |
|
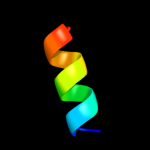
PDB 1onv chain B
Region: 38 - 53
Aligned: 16
Modelled: 16
Confidence: 17.4%
Identity: 44%
PDB header:transcription
Chain: B: PDB Molecule:serine phosphatase fcp1a;
PDBTitle: nmr structure of a complex containing the tfiif subunit2 rap74 and the rnap ii ctd phosphatase fcp1
Phyre2
| 3 |
|
PDB 2kou chain A
Region: 80 - 92
Aligned: 13
Modelled: 13
Confidence: 16.9%
Identity: 15%
PDB header:hydrolase
Chain: A: PDB Molecule:dicer-like protein 4;
PDBTitle: dicer like protein
Phyre2
| 4 |
|
PDB 2xpn chain A
Region: 49 - 89
Aligned: 39
Modelled: 41
Confidence: 15.6%
Identity: 13%
PDB header:transcription
Chain: A: PDB Molecule:iws1;
PDBTitle: crystal structure of a spt6-iws1(spn1) complex from2 encephalitozoon cuniculi, form i
Phyre2
| 5 |
|
PDB 1in0 chain A domain 2
Region: 17 - 32
Aligned: 16
Modelled: 16
Confidence: 11.1%
Identity: 19%
Fold: Ferredoxin-like
Superfamily: YajQ-like
Family: YajQ-like
Phyre2
| 6 |
|
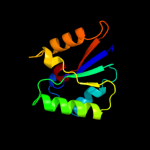
PDB 3h05 chain A
Region: 1 - 103
Aligned: 100
Modelled: 103
Confidence: 10.4%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein vpa0413;
PDBTitle: the crystal structure of a putative nicotinate-nucleotide2 adenylyltransferase from vibrio parahaemolyticus
Phyre2
| 7 |
|
PDB 1wdd chain A domain 2
Region: 16 - 56
Aligned: 41
Modelled: 41
Confidence: 8.4%
Identity: 15%
Fold: Ferredoxin-like
Superfamily: RuBisCO, large subunit, small (N-terminal) domain
Family: Ribulose 1,5-bisphosphate carboxylase-oxygenase
Phyre2
| 8 |
|
PDB 2a7o chain A
Region: 61 - 101
Aligned: 41
Modelled: 41
Confidence: 8.1%
Identity: 27%
PDB header:transcription
Chain: A: PDB Molecule:huntingtin interacting protein b;
PDBTitle: solution structure of the hset2/hypb sri domain
Phyre2
| 9 |
|
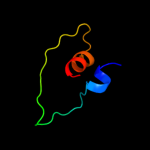
PDB 2r7g chain A domain 1
Region: 69 - 86
Aligned: 18
Modelled: 18
Confidence: 7.6%
Identity: 17%
Fold: Cyclin-like
Superfamily: Cyclin-like
Family: Retinoblastoma tumor suppressor domains
Phyre2
| 10 |
|
PDB 5mdh chain A domain 1
Region: 63 - 109
Aligned: 47
Modelled: 47
Confidence: 6.3%
Identity: 11%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: LDH N-terminal domain-like
Phyre2
| 11 |
|
PDB 7mdh chain A domain 1
Region: 63 - 109
Aligned: 47
Modelled: 47
Confidence: 5.6%
Identity: 21%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: LDH N-terminal domain-like
Phyre2
| 12 |
|
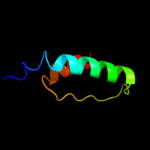
PDB 1q90 chain G
Region: 7 - 18
Aligned: 12
Modelled: 12
Confidence: 5.6%
Identity: 50%
PDB header:photosynthesis
Chain: G: PDB Molecule:cytochrome b6f complex subunit petg;
PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
Phyre2
| 13 |
|
PDB 1q90 chain G
Region: 7 - 18
Aligned: 12
Modelled: 12
Confidence: 5.6%
Identity: 50%
Fold: Single transmembrane helix
Superfamily: PetG subunit of the cytochrome b6f complex
Family: PetG subunit of the cytochrome b6f complex
Phyre2