| 1 |
|
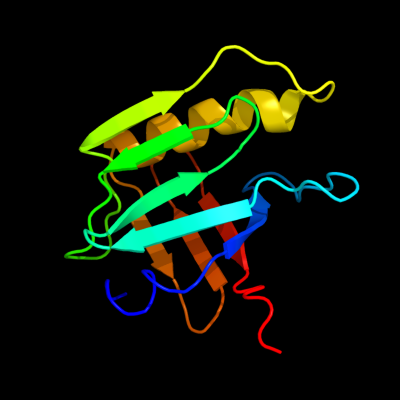
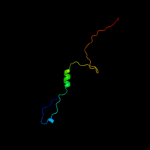
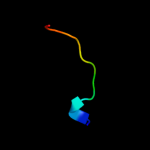
PDB 2kts chain A
Region: 25 - 140
Aligned: 116
Modelled: 116
Confidence: 100.0%
Identity: 100%
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein hslj;
PDBTitle: nmr structure of the protein np_415897.1
Phyre2
| 2 |
|
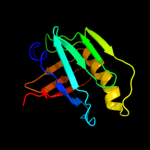
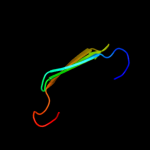
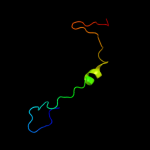
PDB 2la7 chain A
Region: 18 - 136
Aligned: 118
Modelled: 119
Confidence: 99.9%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: nmr structure of the protein yp_557733.1 from burkholderia xenovorans
Phyre2
| 3 |
|
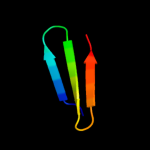
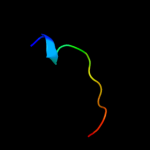
PDB 2y6t chain E
Region: 44 - 96
Aligned: 52
Modelled: 53
Confidence: 83.4%
Identity: 23%
PDB header:hydrolase/inhibitor
Chain: E: PDB Molecule:ecotin;
PDBTitle: molecular recognition of chymotrypsin by the serine2 protease inhibitor ecotin from yersinia pestis
Phyre2
| 4 |
|
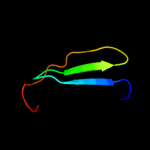
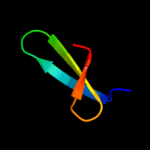
PDB 1ezs chain A
Region: 44 - 96
Aligned: 52
Modelled: 53
Confidence: 81.9%
Identity: 21%
Fold: Ecotin, trypsin inhibitor
Superfamily: Ecotin, trypsin inhibitor
Family: Ecotin, trypsin inhibitor
Phyre2
| 5 |
|
PDB 1xxf chain C
Region: 44 - 96
Aligned: 52
Modelled: 53
Confidence: 78.5%
Identity: 21%
Fold: Ecotin, trypsin inhibitor
Superfamily: Ecotin, trypsin inhibitor
Family: Ecotin, trypsin inhibitor
Phyre2
| 6 |
|
PDB 1slu chain A
Region: 44 - 95
Aligned: 51
Modelled: 52
Confidence: 72.4%
Identity: 22%
Fold: Ecotin, trypsin inhibitor
Superfamily: Ecotin, trypsin inhibitor
Family: Ecotin, trypsin inhibitor
Phyre2
| 7 |
|
PDB 1svb chain A domain 2
Region: 81 - 138
Aligned: 56
Modelled: 58
Confidence: 25.4%
Identity: 13%
Fold: Viral glycoprotein, central and dimerisation domains
Superfamily: Viral glycoprotein, central and dimerisation domains
Family: Viral glycoprotein, central and dimerisation domains
Phyre2
| 8 |
|
PDB 1urz chain C
Region: 81 - 138
Aligned: 56
Modelled: 58
Confidence: 21.1%
Identity: 13%
PDB header:virus/viral protein
Chain: C: PDB Molecule:envelope protein;
PDBTitle: low ph induced, membrane fusion conformation of the2 envelope protein of tick-borne encephalitis virus
Phyre2
| 9 |
|
PDB 1rl6 chain A domain 1
Region: 109 - 140
Aligned: 32
Modelled: 32
Confidence: 20.7%
Identity: 25%
Fold: Ribosomal protein L6
Superfamily: Ribosomal protein L6
Family: Ribosomal protein L6
Phyre2
| 10 |
|
PDB 3ge2 chain A
Region: 112 - 134
Aligned: 23
Modelled: 23
Confidence: 18.4%
Identity: 9%
PDB header:lipoprotein
Chain: A: PDB Molecule:lipoprotein, putative;
PDBTitle: crystal structure of putative lipoprotein sp_0198 from streptococcus2 pneumoniae
Phyre2
| 11 |
|
PDB 2j01 chain H domain 1
Region: 110 - 140
Aligned: 31
Modelled: 31
Confidence: 17.9%
Identity: 16%
Fold: Ribosomal protein L6
Superfamily: Ribosomal protein L6
Family: Ribosomal protein L6
Phyre2
| 12 |
|
PDB 2hnx chain A domain 1
Region: 67 - 79
Aligned: 13
Modelled: 13
Confidence: 15.3%
Identity: 38%
Fold: Lipocalins
Superfamily: Lipocalins
Family: Fatty acid binding protein-like
Phyre2
| 13 |
|
PDB 2of6 chain C
Region: 81 - 127
Aligned: 45
Modelled: 47
Confidence: 13.6%
Identity: 13%
PDB header:virus
Chain: C: PDB Molecule:envelope glycoprotein e;
PDBTitle: structure of immature west nile virus
Phyre2
| 14 |
|
PDB 2q9s chain A
Region: 67 - 79
Aligned: 13
Modelled: 13
Confidence: 13.2%
Identity: 38%
PDB header:lipid binding protein
Chain: A: PDB Molecule:fatty acid-binding protein;
PDBTitle: linoleic acid bound to fatty acid binding protein 4
Phyre2
| 15 |
|
PDB 1jiw chain I
Region: 111 - 133
Aligned: 23
Modelled: 23
Confidence: 11.6%
Identity: 22%
Fold: Streptavidin-like
Superfamily: beta-Barrel protease inhibitors
Family: Metalloprotease inhibitor
Phyre2
| 16 |
|
PDB 2ot9 chain A domain 1
Region: 114 - 139
Aligned: 26
Modelled: 26
Confidence: 9.6%
Identity: 12%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: YaeQ-like
Phyre2
| 17 |
|
PDB 1p58 chain C
Region: 81 - 136
Aligned: 51
Modelled: 56
Confidence: 9.1%
Identity: 18%
PDB header:virus
Chain: C: PDB Molecule:major envelope protein e;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
Phyre2
| 18 |
|
PDB 3c0u chain A
Region: 114 - 138
Aligned: 25
Modelled: 25
Confidence: 8.1%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yaeq;
PDBTitle: crystal structure of e.coli yaeq protein
Phyre2
| 19 |
|
PDB 1ok8 chain A domain 2
Region: 81 - 127
Aligned: 44
Modelled: 47
Confidence: 7.8%
Identity: 16%
Fold: Viral glycoprotein, central and dimerisation domains
Superfamily: Viral glycoprotein, central and dimerisation domains
Family: Viral glycoprotein, central and dimerisation domains
Phyre2
| 20 |
|
PDB 2zjr chain E domain 2
Region: 109 - 140
Aligned: 32
Modelled: 32
Confidence: 7.2%
Identity: 16%
Fold: Ribosomal protein L6
Superfamily: Ribosomal protein L6
Family: Ribosomal protein L6
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|
| 25 |
|