| 1 |
|
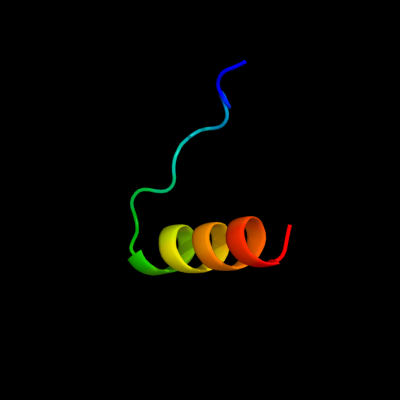
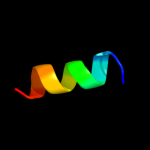
PDB 1vp7 chain B
Region: 19 - 44
Aligned: 26
Modelled: 24
Confidence: 55.7%
Identity: 27%
Fold: Spectrin repeat-like
Superfamily: XseB-like
Family: XseB-like
Phyre2
| 2 |
|
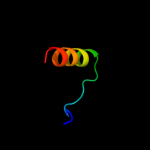
PDB 1vp7 chain A
Region: 29 - 44
Aligned: 16
Modelled: 14
Confidence: 35.2%
Identity: 44%
Fold: Spectrin repeat-like
Superfamily: XseB-like
Family: XseB-like
Phyre2
| 3 |
|
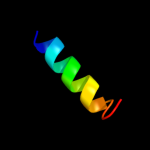
PDB 1vp7 chain D
Region: 29 - 49
Aligned: 21
Modelled: 21
Confidence: 28.0%
Identity: 38%
PDB header:hydrolase
Chain: D: PDB Molecule:exodeoxyribonuclease vii small subunit;
PDBTitle: crystal structure of exodeoxyribonuclease vii small subunit2 (np_881400.1) from bordetella pertussis at 2.40 a resolution
Phyre2
| 4 |
|
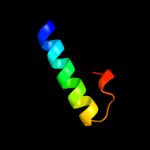
PDB 2ca1 chain A domain 1
Region: 1 - 35
Aligned: 34
Modelled: 35
Confidence: 23.6%
Identity: 24%
Fold: Nucleocapsid protein dimerization domain
Superfamily: Nucleocapsid protein dimerization domain
Family: Coronavirus nucleocapsid protein
Phyre2
| 5 |
|
PDB 2cjr chain A domain 1
Region: 1 - 35
Aligned: 35
Modelled: 35
Confidence: 21.9%
Identity: 31%
Fold: Nucleocapsid protein dimerization domain
Superfamily: Nucleocapsid protein dimerization domain
Family: Coronavirus nucleocapsid protein
Phyre2
| 6 |
|
PDB 3ayh chain A
Region: 33 - 51
Aligned: 19
Modelled: 19
Confidence: 16.4%
Identity: 32%
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase iii subunit rpc9;
PDBTitle: crystal structure of the c17/25 subcomplex from s. pombe rna2 polymerase iii
Phyre2
| 7 |
|
PDB 2omd chain B
Region: 29 - 46
Aligned: 18
Modelled: 18
Confidence: 10.9%
Identity: 33%
PDB header:lyase
Chain: B: PDB Molecule:molybdopterin-converting factor subunit 2;
PDBTitle: crystal structure of molybdopterin converting factor subunit 22 (aq_2181) from aquifex aeolicus vf5
Phyre2
| 8 |
|
PDB 1ni8 chain A
Region: 30 - 42
Aligned: 13
Modelled: 13
Confidence: 9.4%
Identity: 54%
Fold: H-NS histone-like proteins
Superfamily: H-NS histone-like proteins
Family: H-NS histone-like proteins
Phyre2
| 9 |
|
PDB 2kkr chain A
Region: 21 - 38
Aligned: 18
Modelled: 18
Confidence: 8.8%
Identity: 17%
PDB header:transcription, protein binding
Chain: A: PDB Molecule:ataxin-7;
PDBTitle: solution structure of sca7 zinc finger domain from human ataxin-72 protein
Phyre2
| 10 |
|
PDB 3bux chain B domain 2
Region: 26 - 43
Aligned: 18
Modelled: 18
Confidence: 7.2%
Identity: 28%
Fold: N-cbl like
Superfamily: N-terminal domain of cbl (N-cbl)
Family: N-terminal domain of cbl (N-cbl)
Phyre2
| 11 |
|
PDB 1rr7 chain A
Region: 29 - 54
Aligned: 26
Modelled: 26
Confidence: 7.0%
Identity: 27%
PDB header:transcription
Chain: A: PDB Molecule:middle operon regulator;
PDBTitle: crystal structure of the middle operon regulator protein of2 bacteriophage mu
Phyre2
| 12 |
|
PDB 1rr7 chain A
Region: 29 - 54
Aligned: 26
Modelled: 26
Confidence: 7.0%
Identity: 27%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Middle operon regulator, Mor
Phyre2
| 13 |
|
PDB 1oqe chain N
Region: 12 - 19
Aligned: 8
Modelled: 8
Confidence: 6.9%
Identity: 38%
Fold: TNF receptor-like
Superfamily: TNF receptor-like
Family: BAFF receptor-like
Phyre2
| 14 |
|
PDB 1lr1 chain A
Region: 30 - 42
Aligned: 13
Modelled: 13
Confidence: 6.4%
Identity: 54%
Fold: H-NS histone-like proteins
Superfamily: H-NS histone-like proteins
Family: H-NS histone-like proteins
Phyre2