| 1 |
|
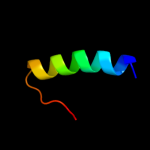
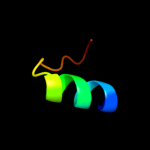
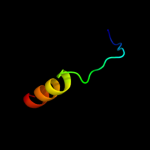
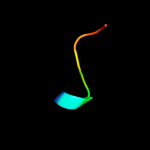
PDB 2rbd chain B
Region: 4 - 27
Aligned: 24
Modelled: 24
Confidence: 25.9%
Identity: 21%
PDB header:structural protein
Chain: B: PDB Molecule:bh2358 protein;
PDBTitle: crystal structure of a putative spore coat protein (bh2358) from2 bacillus halodurans c-125 at 1.54 a resolution
Phyre2
| 2 |
|
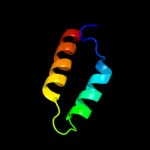
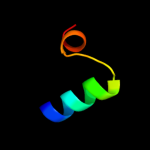
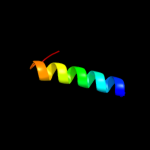
PDB 3d8l chain B
Region: 1 - 41
Aligned: 41
Modelled: 41
Confidence: 22.8%
Identity: 20%
PDB header:viral protein
Chain: B: PDB Molecule:orf12;
PDBTitle: crystal structure of orf12 from the lactococcus lactis bacteriophage2 p2
Phyre2
| 3 |
|
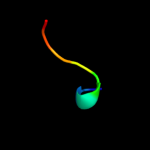
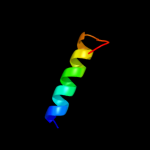
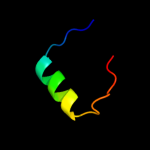
PDB 1uer chain A domain 1
Region: 28 - 35
Aligned: 8
Modelled: 8
Confidence: 18.6%
Identity: 25%
Fold: Long alpha-hairpin
Superfamily: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Phyre2
| 4 |
|
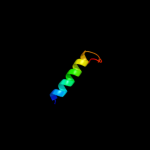
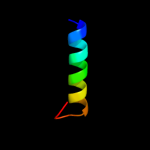
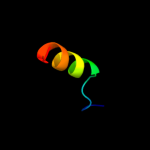
PDB 1uz3 chain A domain 1
Region: 148 - 173
Aligned: 26
Modelled: 26
Confidence: 13.1%
Identity: 12%
Fold: ENT-like
Superfamily: ENT-like
Family: Emsy N terminal (ENT) domain-like
Phyre2
| 5 |
|
PDB 1zke chain A domain 1
Region: 6 - 19
Aligned: 14
Modelled: 14
Confidence: 12.0%
Identity: 43%
Fold: ROP-like
Superfamily: HP1531-like
Family: HP1531-like
Phyre2
| 6 |
|
PDB 3iz5 chain T
Region: 11 - 29
Aligned: 19
Modelled: 19
Confidence: 11.5%
Identity: 16%
PDB header:ribosome
Chain: T: PDB Molecule:60s ribosomal protein l19 (l19e);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
Phyre2
| 7 |
|
PDB 2xoz chain B
Region: 10 - 30
Aligned: 21
Modelled: 21
Confidence: 10.1%
Identity: 14%
PDB header:ligase
Chain: B: PDB Molecule:e3 ubiquitin-protein ligase chfr;
PDBTitle: c-terminal cysteine rich domain of human chfr bound to amp
Phyre2
| 8 |
|
PDB 1p1j chain A domain 1
Region: 150 - 172
Aligned: 23
Modelled: 23
Confidence: 9.4%
Identity: 13%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 9 |
|
PDB 1vko chain A domain 1
Region: 150 - 172
Aligned: 23
Modelled: 23
Confidence: 8.6%
Identity: 13%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 10 |
|
PDB 1vko chain A
Region: 150 - 172
Aligned: 23
Modelled: 23
Confidence: 8.5%
Identity: 13%
PDB header:isomerase
Chain: A: PDB Molecule:inositol-3-phosphate synthase;
PDBTitle: crystal structure of inositol-3-phosphate synthase (ce21227) from2 caenorhabditis elegans at 2.30 a resolution
Phyre2
| 11 |
|
PDB 1hcu chain A
Region: 20 - 44
Aligned: 25
Modelled: 25
Confidence: 8.3%
Identity: 20%
Fold: alpha/alpha toroid
Superfamily: Seven-hairpin glycosidases
Family: Class I alpha-1;2-mannosidase, catalytic domain
Phyre2
| 12 |
|
PDB 1p1h chain D
Region: 150 - 172
Aligned: 23
Modelled: 23
Confidence: 8.0%
Identity: 13%
PDB header:isomerase
Chain: D: PDB Molecule:inositol-3-phosphate synthase;
PDBTitle: crystal structure of the 1l-myo-inositol/nad+ complex
Phyre2
| 13 |
|
PDB 1ma1 chain A domain 1
Region: 28 - 35
Aligned: 8
Modelled: 8
Confidence: 7.7%
Identity: 25%
Fold: Long alpha-hairpin
Superfamily: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Phyre2
| 14 |
|
PDB 1dxs chain A
Region: 86 - 95
Aligned: 10
Modelled: 10
Confidence: 7.0%
Identity: 30%
Fold: SAM domain-like
Superfamily: SAM/Pointed domain
Family: SAM (sterile alpha motif) domain
Phyre2
| 15 |
|
PDB 1b77 chain A domain 1
Region: 147 - 170
Aligned: 24
Modelled: 24
Confidence: 7.0%
Identity: 13%
Fold: DNA clamp
Superfamily: DNA clamp
Family: DNA polymerase processivity factor
Phyre2
| 16 |
|
PDB 2pqr chain D
Region: 22 - 39
Aligned: 18
Modelled: 18
Confidence: 6.9%
Identity: 33%
PDB header:apoptosis
Chain: D: PDB Molecule:wd repeat protein ykr036c;
PDBTitle: crystal structure of yeast fis1 complexed with a fragment of yeast2 caf4
Phyre2
| 17 |
|
PDB 3izc chain T
Region: 12 - 29
Aligned: 18
Modelled: 18
Confidence: 6.6%
Identity: 17%
PDB header:ribosome
Chain: T: PDB Molecule:60s ribosomal protein rpl19 (l19e);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
Phyre2
| 18 |
|
PDB 1n0f chain F
Region: 86 - 95
Aligned: 10
Modelled: 10
Confidence: 6.1%
Identity: 30%
PDB header:biosynthetic protein
Chain: F: PDB Molecule:protein mraz;
PDBTitle: crystal structure of a cell division and cell wall2 biosynthesis protein upf0040 from mycoplasma pneumoniae:3 indication of a novel fold with a possible new conserved4 sequence motif
Phyre2
| 19 |
|
PDB 1bsm chain A domain 1
Region: 28 - 36
Aligned: 9
Modelled: 9
Confidence: 5.6%
Identity: 22%
Fold: Long alpha-hairpin
Superfamily: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Family: Fe,Mn superoxide dismutase (SOD), N-terminal domain
Phyre2
| 20 |
|
PDB 2a5y chain B domain 1
Region: 22 - 34
Aligned: 13
Modelled: 13
Confidence: 5.6%
Identity: 38%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: CED-4 C-terminal domain-like
Phyre2
| 21 |
|