| 1 |
|
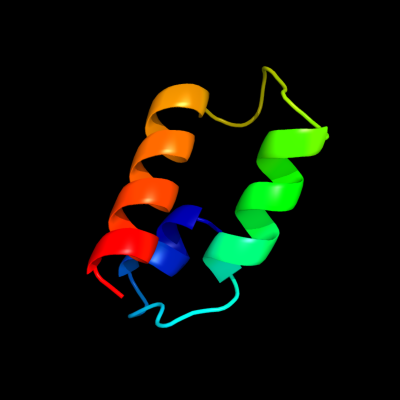
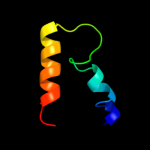
PDB 3ha4 chain C
Region: 19 - 66
Aligned: 48
Modelled: 48
Confidence: 23.2%
Identity: 27%
PDB header:unknown function
Chain: C: PDB Molecule:mix1;
PDBTitle: crystal structure of the type one membrane protein mix1 from2 leishmania
Phyre2
| 2 |
|
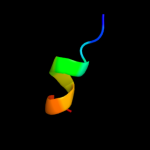
PDB 2ki9 chain A
Region: 35 - 43
Aligned: 9
Modelled: 9
Confidence: 22.4%
Identity: 67%
PDB header:membrane protein
Chain: A: PDB Molecule:cannabinoid receptor 2;
PDBTitle: human cannabinoid receptor-2 helix 6
Phyre2
| 3 |
|
PDB 3kf8 chain C
Region: 22 - 38
Aligned: 17
Modelled: 17
Confidence: 14.4%
Identity: 41%
PDB header:structural protein
Chain: C: PDB Molecule:protein stn1;
PDBTitle: crystal structure of c. tropicalis stn1-ten1 complex
Phyre2
| 4 |
|
PDB 1lva chain A domain 4
Region: 40 - 73
Aligned: 34
Modelled: 34
Confidence: 8.9%
Identity: 21%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: C-terminal fragment of elongation factor SelB
Phyre2
| 5 |
|
PDB 3hm5 chain A
Region: 47 - 57
Aligned: 11
Modelled: 11
Confidence: 8.8%
Identity: 27%
PDB header:transcription
Chain: A: PDB Molecule:dna methyltransferase 1-associated protein 1;
PDBTitle: sant domain of human dna methyltransferase 1 associated2 protein 1
Phyre2
| 6 |
|
PDB 1w7p chain D domain 2
Region: 51 - 59
Aligned: 9
Modelled: 9
Confidence: 8.0%
Identity: 11%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Vacuolar sorting protein domain
Phyre2
| 7 |
|
PDB 1pd0 chain A domain 1
Region: 31 - 68
Aligned: 37
Modelled: 38
Confidence: 7.5%
Identity: 22%
Fold: ERP29 C domain-like
Superfamily: Helical domain of Sec23/24
Family: Helical domain of Sec23/24
Phyre2
| 8 |
|
PDB 1nlx chain A
Region: 35 - 54
Aligned: 18
Modelled: 20
Confidence: 6.3%
Identity: 33%
Fold: Four-helical up-and-down bundle
Superfamily: Group V grass pollen allergen
Family: Group V grass pollen allergen
Phyre2
| 9 |
|
PDB 1jdh chain B
Region: 46 - 56
Aligned: 11
Modelled: 11
Confidence: 6.1%
Identity: 18%
PDB header:transcription
Chain: B: PDB Molecule:htcf-4;
PDBTitle: crystal structure of beta-catenin and htcf-4
Phyre2
| 10 |
|
PDB 1ckx chain A
Region: 52 - 63
Aligned: 12
Modelled: 12
Confidence: 5.8%
Identity: 25%
PDB header:metal transport
Chain: A: PDB Molecule:cystic fibrosis transmembrane conductance
PDBTitle: cystic fibrosis transmembrane conductance regulator:2 solution structures of peptides based on the phe508 region,3 the most common site of disease-causing delta-f508 mutation
Phyre2