| 1 |
|
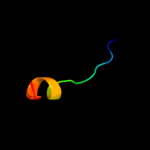
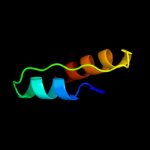
PDB 1v1h chain A domain 1
Region: 27 - 44
Aligned: 18
Modelled: 18
Confidence: 16.3%
Identity: 44%
Fold: Triple beta-spiral
Superfamily: Fibre shaft of virus attachment proteins
Family: Adenovirus
Phyre2
| 2 |
|
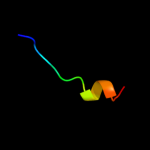
PDB 2zif chain B
Region: 38 - 75
Aligned: 38
Modelled: 38
Confidence: 14.8%
Identity: 21%
PDB header:transferase
Chain: B: PDB Molecule:putative modification methylase;
PDBTitle: crystal structure of ttha0409, putative dna modification2 methylase from thermus thermophilus hb8- complexed with s-3 adenosyl-l-methionine
Phyre2
| 3 |
|
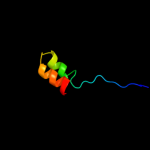
PDB 1o59 chain A domain 2
Region: 10 - 38
Aligned: 29
Modelled: 29
Confidence: 13.7%
Identity: 24%
Fold: Galactose-binding domain-like
Superfamily: Galactose-binding domain-like
Family: Allantoicase repeat
Phyre2
| 4 |
|
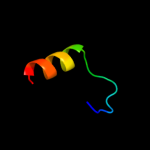
PDB 1sg3 chain A
Region: 10 - 38
Aligned: 29
Modelled: 29
Confidence: 11.3%
Identity: 24%
PDB header:hydrolase
Chain: A: PDB Molecule:allantoicase;
PDBTitle: structure of allantoicase
Phyre2
| 5 |
|
PDB 1g60 chain A
Region: 38 - 73
Aligned: 36
Modelled: 36
Confidence: 10.4%
Identity: 25%
Fold: S-adenosyl-L-methionine-dependent methyltransferases
Superfamily: S-adenosyl-L-methionine-dependent methyltransferases
Family: Type II DNA methylase
Phyre2
| 6 |
|
PDB 1n6j chain G
Region: 66 - 80
Aligned: 15
Modelled: 15
Confidence: 10.0%
Identity: 73%
PDB header:transcription/dna
Chain: G: PDB Molecule:calcineurin-binding protein cabin 1;
PDBTitle: structural basis of sequence-specific recruitment of2 histone deacetylases by myocyte enhancer factor-2
Phyre2
| 7 |
|
PDB 191l chain A
Region: 37 - 142
Aligned: 106
Modelled: 106
Confidence: 8.4%
Identity: 15%
Fold: Lysozyme-like
Superfamily: Lysozyme-like
Family: Phage lysozyme
Phyre2
| 8 |
|
PDB 1js8 chain A domain 2
Region: 80 - 93
Aligned: 14
Modelled: 14
Confidence: 8.0%
Identity: 29%
Fold: C-terminal domain of mollusc hemocyanin
Superfamily: C-terminal domain of mollusc hemocyanin
Family: C-terminal domain of mollusc hemocyanin
Phyre2
| 9 |
|
PDB 1lnl chain A domain 2
Region: 80 - 93
Aligned: 14
Modelled: 14
Confidence: 7.7%
Identity: 50%
Fold: C-terminal domain of mollusc hemocyanin
Superfamily: C-terminal domain of mollusc hemocyanin
Family: C-terminal domain of mollusc hemocyanin
Phyre2
| 10 |
|
PDB 1o59 chain A domain 1
Region: 10 - 38
Aligned: 29
Modelled: 28
Confidence: 7.6%
Identity: 10%
Fold: Galactose-binding domain-like
Superfamily: Galactose-binding domain-like
Family: Allantoicase repeat
Phyre2
| 11 |
|
PDB 1r48 chain A
Region: 62 - 70
Aligned: 9
Modelled: 9
Confidence: 6.9%
Identity: 67%
PDB header:transport protein
Chain: A: PDB Molecule:proline/betaine transporter;
PDBTitle: solution structure of the c-terminal cytoplasmic domain2 residues 468-497 of escherichia coli protein prop
Phyre2
| 12 |
|
PDB 2i2x chain O
Region: 41 - 76
Aligned: 35
Modelled: 36
Confidence: 6.6%
Identity: 29%
PDB header:transferase
Chain: O: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
Phyre2
| 13 |
|
PDB 1e3h chain A domain 6
Region: 59 - 78
Aligned: 20
Modelled: 20
Confidence: 6.1%
Identity: 25%
Fold: Ribonuclease PH domain 2-like
Superfamily: Ribonuclease PH domain 2-like
Family: Ribonuclease PH domain 2-like
Phyre2
| 14 |
|
PDB 1nw6 chain A
Region: 38 - 74
Aligned: 37
Modelled: 37
Confidence: 5.9%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:modification methylase rsri;
PDBTitle: structure of the beta class n6-adenine dna methyltransferase rsri2 bound to sinefungin
Phyre2
| 15 |
|
PDB 2iec chain A domain 1
Region: 4 - 63
Aligned: 49
Modelled: 60
Confidence: 5.7%
Identity: 22%
Fold: MK0786-like
Superfamily: MK0786-like
Family: MK0786-like
Phyre2
| 16 |
|
PDB 2v6z chain M
Region: 69 - 114
Aligned: 46
Modelled: 46
Confidence: 5.4%
Identity: 17%
PDB header:transferase
Chain: M: PDB Molecule:dna polymerase epsilon subunit 2;
PDBTitle: solution structure of amino terminal domain of human dna2 polymerase epsilon subunit b
Phyre2