| 1 |
|
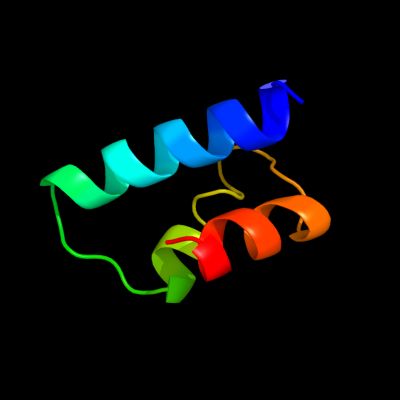
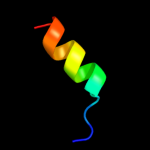
PDB 3e3v chain A
Region: 14 - 59
Aligned: 46
Modelled: 46
Confidence: 14.6%
Identity: 17%
PDB header:recombination
Chain: A: PDB Molecule:regulatory protein recx;
PDBTitle: crystal structure of recx from lactobacillus salivarius
Phyre2
| 2 |
|
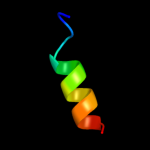
PDB 3se4 chain B
Region: 45 - 59
Aligned: 15
Modelled: 15
Confidence: 10.1%
Identity: 27%
PDB header:immune system receptor
Chain: B: PDB Molecule:interferon omega-1;
PDBTitle: human ifnw-ifnar ternary complex
Phyre2
| 3 |
|
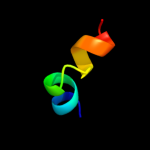
PDB 3oq3 chain A
Region: 45 - 59
Aligned: 15
Modelled: 15
Confidence: 9.9%
Identity: 27%
PDB header:cytokine/viral protein
Chain: A: PDB Molecule:interferon alpha-5;
PDBTitle: structural basis of type-i interferon sequestration by a poxvirus2 decoy receptor
Phyre2
| 4 |
|
PDB 1au1 chain A
Region: 45 - 59
Aligned: 15
Modelled: 15
Confidence: 9.6%
Identity: 13%
Fold: 4-helical cytokines
Superfamily: 4-helical cytokines
Family: Interferons/interleukin-10 (IL-10)
Phyre2
| 5 |
|
PDB 3piw chain A
Region: 45 - 59
Aligned: 15
Modelled: 15
Confidence: 9.6%
Identity: 7%
PDB header:cytokine
Chain: A: PDB Molecule:type i interferon 2;
PDBTitle: zebrafish interferon 2
Phyre2
| 6 |
|
PDB 1rh2 chain A
Region: 45 - 59
Aligned: 15
Modelled: 15
Confidence: 9.5%
Identity: 27%
Fold: 4-helical cytokines
Superfamily: 4-helical cytokines
Family: Interferons/interleukin-10 (IL-10)
Phyre2
| 7 |
|
PDB 1ogl chain A
Region: 18 - 61
Aligned: 44
Modelled: 44
Confidence: 9.2%
Identity: 18%
Fold: all-alpha NTP pyrophosphatases
Superfamily: all-alpha NTP pyrophosphatases
Family: Type II deoxyuridine triphosphatase
Phyre2
| 8 |
|
PDB 1b5l chain A
Region: 45 - 59
Aligned: 15
Modelled: 15
Confidence: 8.6%
Identity: 33%
Fold: 4-helical cytokines
Superfamily: 4-helical cytokines
Family: Interferons/interleukin-10 (IL-10)
Phyre2
| 9 |
|
PDB 3piv chain A
Region: 45 - 59
Aligned: 15
Modelled: 15
Confidence: 8.3%
Identity: 20%
PDB header:cytokine
Chain: A: PDB Molecule:interferon;
PDBTitle: zebrafish interferon 1
Phyre2
| 10 |
|
PDB 1wu3 chain I
Region: 45 - 59
Aligned: 15
Modelled: 15
Confidence: 8.0%
Identity: 7%
Fold: 4-helical cytokines
Superfamily: 4-helical cytokines
Family: Interferons/interleukin-10 (IL-10)
Phyre2
| 11 |
|
PDB 3fwc chain O
Region: 38 - 53
Aligned: 16
Modelled: 16
Confidence: 6.2%
Identity: 6%
PDB header:cell cycle, transcription
Chain: O: PDB Molecule:protein sus1;
PDBTitle: sac3:sus1:cdc31 complex
Phyre2
| 12 |
|
PDB 2rrh chain A
Region: 50 - 58
Aligned: 9
Modelled: 9
Confidence: 5.7%
Identity: 56%
PDB header:hormone
Chain: A: PDB Molecule:vip peptides;
PDBTitle: nmr structure of vasoactive intestinal peptide in methanol
Phyre2