| 1 |
|
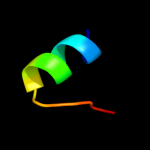
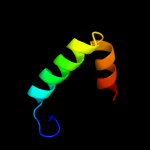
PDB 1m0f chain B
Region: 216 - 229
Aligned: 14
Modelled: 14
Confidence: 31.1%
Identity: 36%
PDB header:virus
Chain: B: PDB Molecule:scaffolding protein b;
PDBTitle: structural studies of bacteriophage alpha3 assembly, cryo-2 electron microscopy
Phyre2
| 2 |
|
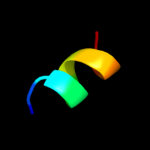
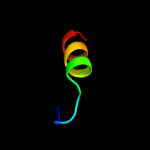
PDB 1pvg chain A domain 1
Region: 77 - 86
Aligned: 10
Modelled: 10
Confidence: 16.1%
Identity: 40%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: DNA gyrase/MutL, second domain
Phyre2
| 3 |
|
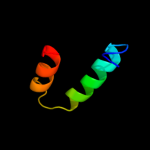
PDB 2v1s chain D
Region: 74 - 107
Aligned: 34
Modelled: 34
Confidence: 15.8%
Identity: 12%
PDB header:oxidoreductase
Chain: D: PDB Molecule:mitochondrial import receptor subunit tom20 homolog;
PDBTitle: crystal structure of rat tom20-aldh presequence complex
Phyre2
| 4 |
|
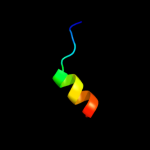
PDB 2d6f chain C domain 1
Region: 96 - 106
Aligned: 11
Modelled: 11
Confidence: 14.5%
Identity: 36%
Fold: GatB/YqeY motif
Superfamily: GatB/YqeY motif
Family: GatB/GatE C-terminal domain-like
Phyre2
| 5 |
|
PDB 1ckq chain A
Region: 92 - 106
Aligned: 15
Modelled: 15
Confidence: 10.4%
Identity: 33%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Restriction endonuclease EcoRI
Phyre2
| 6 |
|
PDB 1om2 chain A
Region: 74 - 107
Aligned: 34
Modelled: 34
Confidence: 10.2%
Identity: 12%
Fold: Open three-helical up-and-down bundle
Superfamily: Mitochondrial import receptor subunit Tom20
Family: Mitochondrial import receptor subunit Tom20
Phyre2
| 7 |
|
PDB 1kij chain A domain 1
Region: 77 - 86
Aligned: 10
Modelled: 10
Confidence: 9.9%
Identity: 40%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: DNA gyrase/MutL, second domain
Phyre2
| 8 |
|
PDB 1s16 chain A domain 1
Region: 77 - 86
Aligned: 10
Modelled: 10
Confidence: 9.7%
Identity: 50%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: DNA gyrase/MutL, second domain
Phyre2
| 9 |
|
PDB 1ei1 chain A domain 1
Region: 77 - 86
Aligned: 10
Modelled: 10
Confidence: 9.6%
Identity: 50%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: DNA gyrase/MutL, second domain
Phyre2
| 10 |
|
PDB 1otf chain A
Region: 91 - 106
Aligned: 16
Modelled: 16
Confidence: 9.4%
Identity: 13%
Fold: Tautomerase/MIF
Superfamily: Tautomerase/MIF
Family: 4-oxalocrotonate tautomerase-like
Phyre2
| 11 |
|
PDB 1bjp chain A
Region: 91 - 106
Aligned: 16
Modelled: 16
Confidence: 6.0%
Identity: 19%
Fold: Tautomerase/MIF
Superfamily: Tautomerase/MIF
Family: 4-oxalocrotonate tautomerase-like
Phyre2