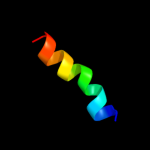
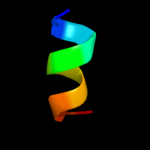| 1 |
|
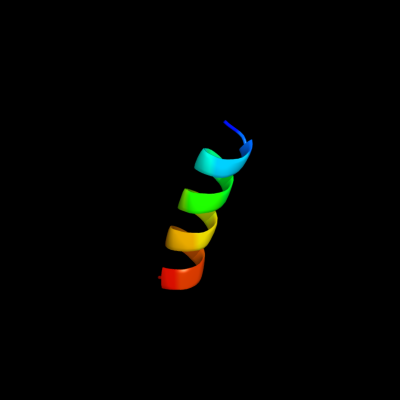
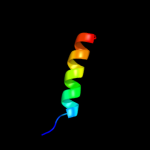
PDB 2l9u chain A
Region: 1 - 17
Aligned: 17
Modelled: 17
Confidence: 38.0%
Identity: 41%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-3;
PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
Phyre2
| 2 |
|
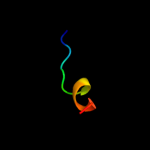
PDB 1i2t chain A
Region: 3 - 11
Aligned: 9
Modelled: 9
Confidence: 20.5%
Identity: 67%
Fold: PABP domain-like
Superfamily: PABC (PABP) domain
Family: PABC (PABP) domain
Phyre2
| 3 |
|
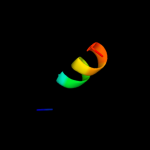
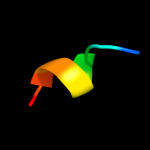
PDB 3pth chain A
Region: 3 - 11
Aligned: 9
Modelled: 9
Confidence: 20.3%
Identity: 78%
PDB header:rna binding protein
Chain: A: PDB Molecule:polyadenylate-binding protein 1;
PDBTitle: the pabc1 mlle domain bound to the variant pam2 motif of larp4b
Phyre2
| 4 |
|
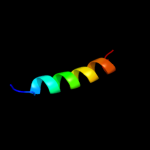
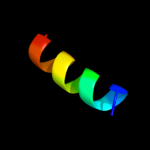
PDB 2dyd chain A
Region: 3 - 13
Aligned: 11
Modelled: 11
Confidence: 20.0%
Identity: 45%
PDB header:rna binding protein
Chain: A: PDB Molecule:poly(a)-binding protein;
PDBTitle: solution structure of the pabc domain from triticum2 aevestium poly(a)-binding protein
Phyre2
| 5 |
|
PDB 1ifw chain A
Region: 3 - 11
Aligned: 9
Modelled: 9
Confidence: 18.8%
Identity: 67%
Fold: PABP domain-like
Superfamily: PABC (PABP) domain
Family: PABC (PABP) domain
Phyre2
| 6 |
|
PDB 1nmr chain A
Region: 3 - 11
Aligned: 9
Modelled: 9
Confidence: 13.3%
Identity: 56%
Fold: PABP domain-like
Superfamily: PABC (PABP) domain
Family: PABC (PABP) domain
Phyre2
| 7 |
|
PDB 2e74 chain G domain 1
Region: 11 - 22
Aligned: 12
Modelled: 12
Confidence: 12.3%
Identity: 67%
Fold: Single transmembrane helix
Superfamily: PetG subunit of the cytochrome b6f complex
Family: PetG subunit of the cytochrome b6f complex
Phyre2
| 8 |
|
PDB 1jgn chain A
Region: 3 - 11
Aligned: 9
Modelled: 9
Confidence: 12.2%
Identity: 78%
Fold: PABP domain-like
Superfamily: PABC (PABP) domain
Family: PABC (PABP) domain
Phyre2
| 9 |
|
PDB 2l35 chain B
Region: 1 - 20
Aligned: 20
Modelled: 20
Confidence: 12.1%
Identity: 35%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
Phyre2
| 10 |
|
PDB 2l2t chain A
Region: 3 - 21
Aligned: 19
Modelled: 19
Confidence: 11.7%
Identity: 53%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
Phyre2
| 11 |
|
PDB 2l34 chain A
Region: 1 - 20
Aligned: 20
Modelled: 20
Confidence: 11.4%
Identity: 35%
PDB header:protein binding
Chain: A: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 12 |
|
PDB 2l34 chain B
Region: 1 - 20
Aligned: 20
Modelled: 20
Confidence: 11.4%
Identity: 35%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 13 |
|
PDB 1vf5 chain G
Region: 11 - 22
Aligned: 12
Modelled: 12
Confidence: 11.1%
Identity: 67%
PDB header:photosynthesis
Chain: G: PDB Molecule:protein pet g;
PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
Phyre2
| 14 |
|
PDB 1vf5 chain G
Region: 11 - 22
Aligned: 12
Modelled: 12
Confidence: 11.1%
Identity: 67%
Fold: Single transmembrane helix
Superfamily: PetG subunit of the cytochrome b6f complex
Family: PetG subunit of the cytochrome b6f complex
Phyre2
| 15 |
|
PDB 2y69 chain Q
Region: 5 - 29
Aligned: 25
Modelled: 25
Confidence: 7.9%
Identity: 12%
PDB header:electron transport
Chain: Q: PDB Molecule:cytochrome c oxidase subunit 4 isoform 1;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 16 |
|
PDB 1g9l chain A
Region: 3 - 11
Aligned: 9
Modelled: 9
Confidence: 7.6%
Identity: 78%
Fold: PABP domain-like
Superfamily: PABC (PABP) domain
Family: PABC (PABP) domain
Phyre2
| 17 |
|
PDB 2k9y chain B
Region: 4 - 17
Aligned: 14
Modelled: 14
Confidence: 6.7%
Identity: 50%
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 18 |
|
PDB 2k9y chain A
Region: 4 - 17
Aligned: 14
Modelled: 14
Confidence: 6.7%
Identity: 50%
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 19 |
|
PDB 2k1k chain B
Region: 5 - 21
Aligned: 17
Modelled: 17
Confidence: 6.2%
Identity: 65%
PDB header:signaling protein
Chain: B: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
Phyre2
| 20 |
|
PDB 2k1l chain A
Region: 5 - 21
Aligned: 17
Modelled: 17
Confidence: 6.2%
Identity: 65%
PDB header:signaling protein
Chain: A: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|