| 1 |
|
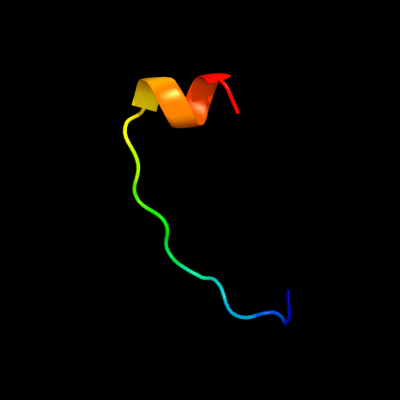
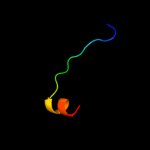
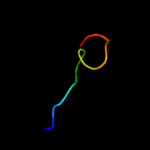
PDB 1bdt chain A
Region: 118 - 135
Aligned: 18
Modelled: 18
Confidence: 20.3%
Identity: 44%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: Arc/Mnt-like phage repressors
Phyre2
| 2 |
|
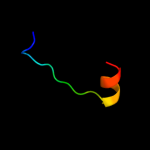
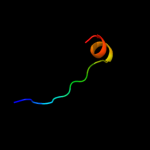
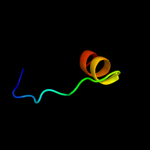
PDB 1b28 chain A
Region: 118 - 135
Aligned: 18
Modelled: 18
Confidence: 19.4%
Identity: 44%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: Arc/Mnt-like phage repressors
Phyre2
| 3 |
|
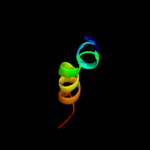
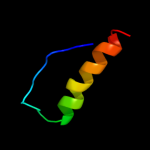
PDB 2wfu chain A
Region: 73 - 94
Aligned: 16
Modelled: 22
Confidence: 14.5%
Identity: 81%
PDB header:signaling protein
Chain: A: PDB Molecule:probable insulin-like peptide 5 a chain;
PDBTitle: crystal structure of dilp5 variant db
Phyre2
| 4 |
|
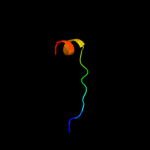
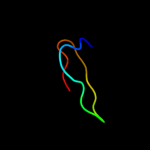
PDB 2wfv chain A
Region: 73 - 94
Aligned: 16
Modelled: 22
Confidence: 14.0%
Identity: 81%
PDB header:signaling protein
Chain: A: PDB Molecule:probable insulin-like peptide 5 a chain;
PDBTitle: crystal structure of dilp5 variant c4
Phyre2
| 5 |
|
PDB 1myl chain A
Region: 119 - 135
Aligned: 17
Modelled: 17
Confidence: 10.0%
Identity: 41%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: Arc/Mnt-like phage repressors
Phyre2
| 6 |
|
PDB 1baz chain A
Region: 118 - 135
Aligned: 18
Modelled: 18
Confidence: 8.8%
Identity: 44%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: Arc/Mnt-like phage repressors
Phyre2
| 7 |
|
PDB 1myk chain A
Region: 119 - 135
Aligned: 17
Modelled: 17
Confidence: 8.3%
Identity: 41%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: Arc/Mnt-like phage repressors
Phyre2
| 8 |
|
PDB 1xhj chain A
Region: 146 - 162
Aligned: 17
Modelled: 17
Confidence: 7.8%
Identity: 35%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Fe-S cluster assembly (FSCA) domain-like
Family: NifU C-terminal domain-like
Phyre2
| 9 |
|
PDB 1veh chain A
Region: 146 - 177
Aligned: 32
Modelled: 32
Confidence: 7.4%
Identity: 22%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Fe-S cluster assembly (FSCA) domain-like
Family: NifU C-terminal domain-like
Phyre2
| 10 |
|
PDB 3i3a chain C
Region: 169 - 188
Aligned: 20
Modelled: 20
Confidence: 6.7%
Identity: 20%
PDB header:transferase
Chain: C: PDB Molecule:acyl-[acyl-carrier-protein]--udp-n-
PDBTitle: structural basis for the sugar nucleotide and acyl chain2 selectivity of leptospira interrogans lpxa
Phyre2
| 11 |
|
PDB 1th5 chain A domain 1
Region: 146 - 160
Aligned: 15
Modelled: 15
Confidence: 6.6%
Identity: 7%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Fe-S cluster assembly (FSCA) domain-like
Family: NifU C-terminal domain-like
Phyre2
| 12 |
|
PDB 1z2t chain A
Region: 28 - 37
Aligned: 10
Modelled: 10
Confidence: 6.4%
Identity: 50%
PDB header:lipid binding protein
Chain: A: PDB Molecule:anchor peptide ser65-leu87 of almgs;
PDBTitle: nmr structure study of anchor peptide ser65-leu87 of enzyme2 acholeplasma laidlawii monoglycosyldiacyl glycerol3 synthase (almgs) in dhpc micelles
Phyre2
| 13 |
|
PDB 2jnv chain A
Region: 146 - 162
Aligned: 17
Modelled: 17
Confidence: 6.3%
Identity: 29%
PDB header:metal transport
Chain: A: PDB Molecule:nifu-like protein 1, chloroplast;
PDBTitle: solution structure of c-terminal domain of nifu-like2 protein from oryza sativa
Phyre2
| 14 |
|
PDB 1n12 chain A
Region: 97 - 128
Aligned: 32
Modelled: 32
Confidence: 6.2%
Identity: 22%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Bacterial adhesins
Family: Pilus subunits
Phyre2
| 15 |
|
PDB 2h8b chain B
Region: 15 - 34
Aligned: 20
Modelled: 20
Confidence: 6.1%
Identity: 40%
PDB header:hormone/growth factor
Chain: B: PDB Molecule:insulin-like 3;
PDBTitle: solution structure of insl3
Phyre2